
Hierarchical clustering
Source:vignettes/articles/hierarchial-clustering.Rmd
hierarchial-clustering.RmdTheory
Traditional hierarchical agglomerative clustering (HAC) is usually carried out in two mandatory steps with a third optional one:
- Compute the distance matrix in which entry stores the distance between observation and observation ;
- Compute the hierarchy of clustering structures in the form of a dendrogram by first assuming each observation belongs to its own cluster and progressively merge observations and groups of observations together using a so-called linkage criterion;
- [optional] Cut the tree at a given distance in order to form clusters.
Each of these steps incurs slight modifications when it comes to clustering functional data.
Step 1. Functional HAC needs to account for the
intrinsic amplitude and phase variability inherent to functional data.
It is therefore natural when computing the distance between two curves
to minimize such a distance over all possible warping functions in a
chosen class, i.e. to integrate alignment when computing the distance
matrix. The idea is to assess how far two curves are after optimal phase
alignment. This step is achieved through the fdadist().
Step 2. The process of building up the hierarchy of
possible clustering structures does not change once the distance matrix
is known. In fact, from a practical point of view with R,
it is still achieved via a call to stats::hclust().
Here, the user can choose which linkage criterion to use for assessing
distances between sets of curves. For the sake of simplicity, only a
subset of choices from the stats::hclust()
function is actually available in fdacluster,
namely complete, average, single
and ward.D2.
Step 3. Once the dendrogram is built, a big
difference between multivariate HAC and functional HAC stems from the
fact that the latter requires the choice of the number of clusters,
hence making Step 3 mandatory as well. This is because, once the
grouping structure has been chosen, all the curves need to undergo a
last alignment to the centroid of the cluster they have been assigned
to. Then, and only then, within-cluster sum of squared distances to
centroid and silhouette values can be assessed. This step is achieved
through a call to fdakmeans()
with
and initial centroid corresponding to the cluster medoid.
Using fdacluster
All these steps have been enpapsulated into a single function in fdacluster for ease of usage:
fdahclust(
x,
y,
n_clusters = 1L,
is_domain_interval = FALSE,
transformation = c("identity", "srvf"),
warping_class = c("none", "shift", "dilation", "affine", "bpd"),
centroid_type = c("mean", "median", "medoid", "lowess", "poly"),
metric = c("l2", "normalized_l2", "pearson"),
linkage_criterion = c("complete", "average", "single", "ward.D2"),
cluster_on_phase = FALSE,
use_verbose = TRUE,
warping_options = c(0.15, 0.15),
maximum_number_of_iterations = 100L,
number_of_threads = 1L,
parallel_method = 0L,
distance_relative_tolerance = 0.001,
use_fence = FALSE,
check_total_dissimilarity = TRUE,
compute_overall_center = FALSE
)The first group of arguments in the above call are the arguments that
the user is the most likely to interact with. The number of clusters can
be specified through the argument n_clusters, the choice of
the class of warping functions or type of centroid or metric is achieved
through the arguments warping_class,
centroid_type and metric respectively. The
linkage criterion can be selected via linkage_criterion. By
default, the function performs affine registration using the pointwise
sample mean for computing cluster centroid with the L2 distance between
original curves and complete linkage. By default, the function uses
amplitude data after alignment for assigning cluster membership of the
observed curves. The user can switch on the use of phase data instead by
using cluster_on_phase = TRUE.
The second group of arguments are set to sensible default values and
are used for the internal call to fdakmeans()
with
,
which performs within-cluster alignment to centroid once groups have
been determined by cutting the dendrogram at the appropriate height.
These arguments are not meant to be changed unless the results of the
clustering via fdahclust()
seem very odd.
Example
Simulated data set
Let us consider the following sample of
simulated unidimensional curves:
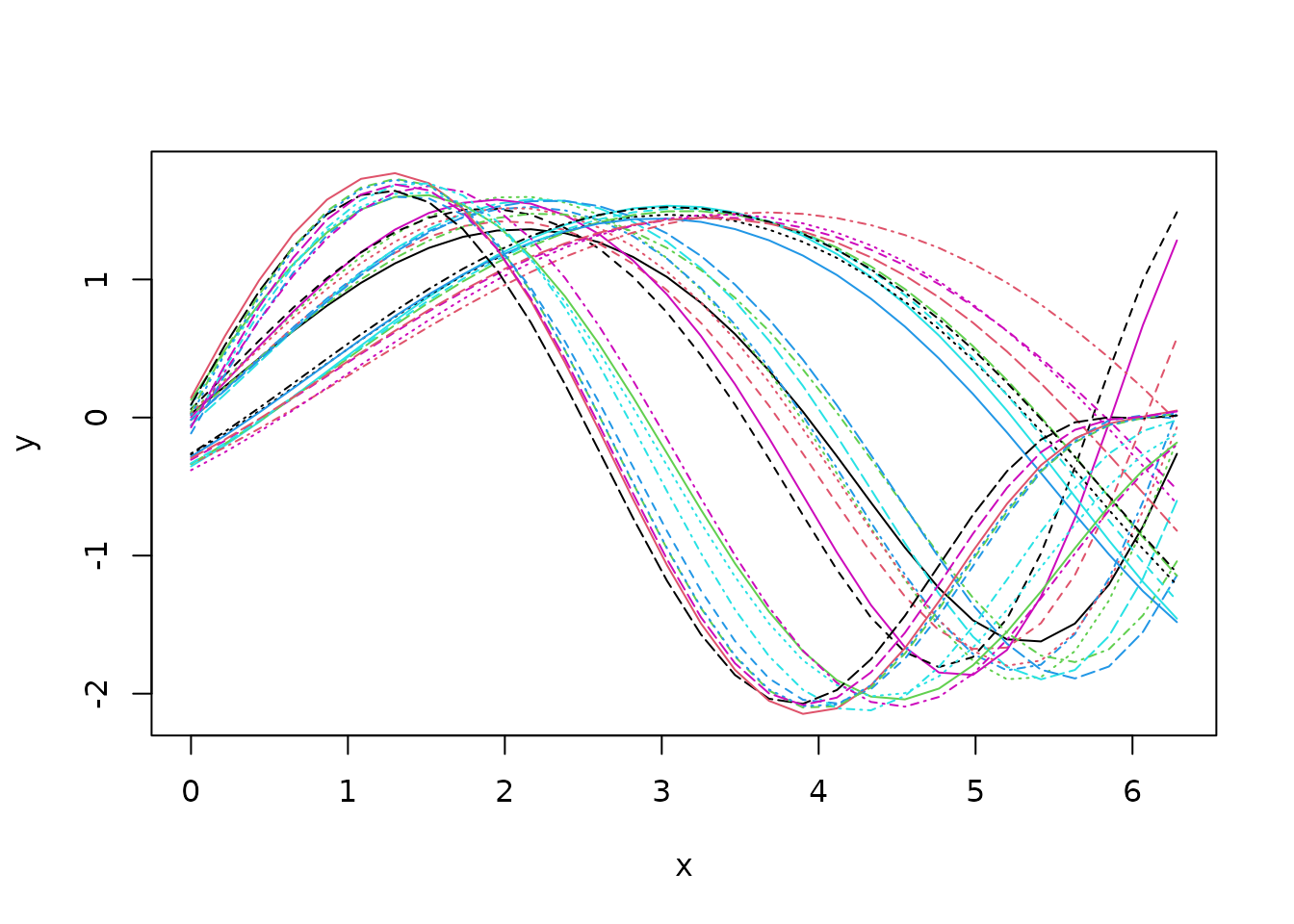
Looking at the data set, it seems that we shall expect groups if we aim at clustering based on phase variability but probably only groups if we search for a clustering structure based on amplitude variability.
HAC based on amplitude variability
We can perform HAC based on amplitude variability as follows:
out1 <- fdahclust(
simulated30_sub$x,
simulated30_sub$y,
n_clusters = 2,
centroid_type = "mean",
warping_class = "affine",
metric = "normalized_l2",
cluster_on_phase = FALSE
)The fdahclust()
function returns an object of class caps
(for Clustering with Amplitude and
Phase Separation) for which
S3 specialized methods of ggplot2::autoplot()
and graphics::plot()
have been implemented.
For instance, we can visualize the original and aligned functional data set with:
plot(out1, type = "amplitude")
#> Warning in data.frame(grid = as.vector(grids), value = as.vector(unicurves), :
#> row names were found from a short variable and have been discarded
#> Warning in data.frame(grid = as.vector(grids), value = as.vector(unicurves), :
#> row names were found from a short variable and have been discarded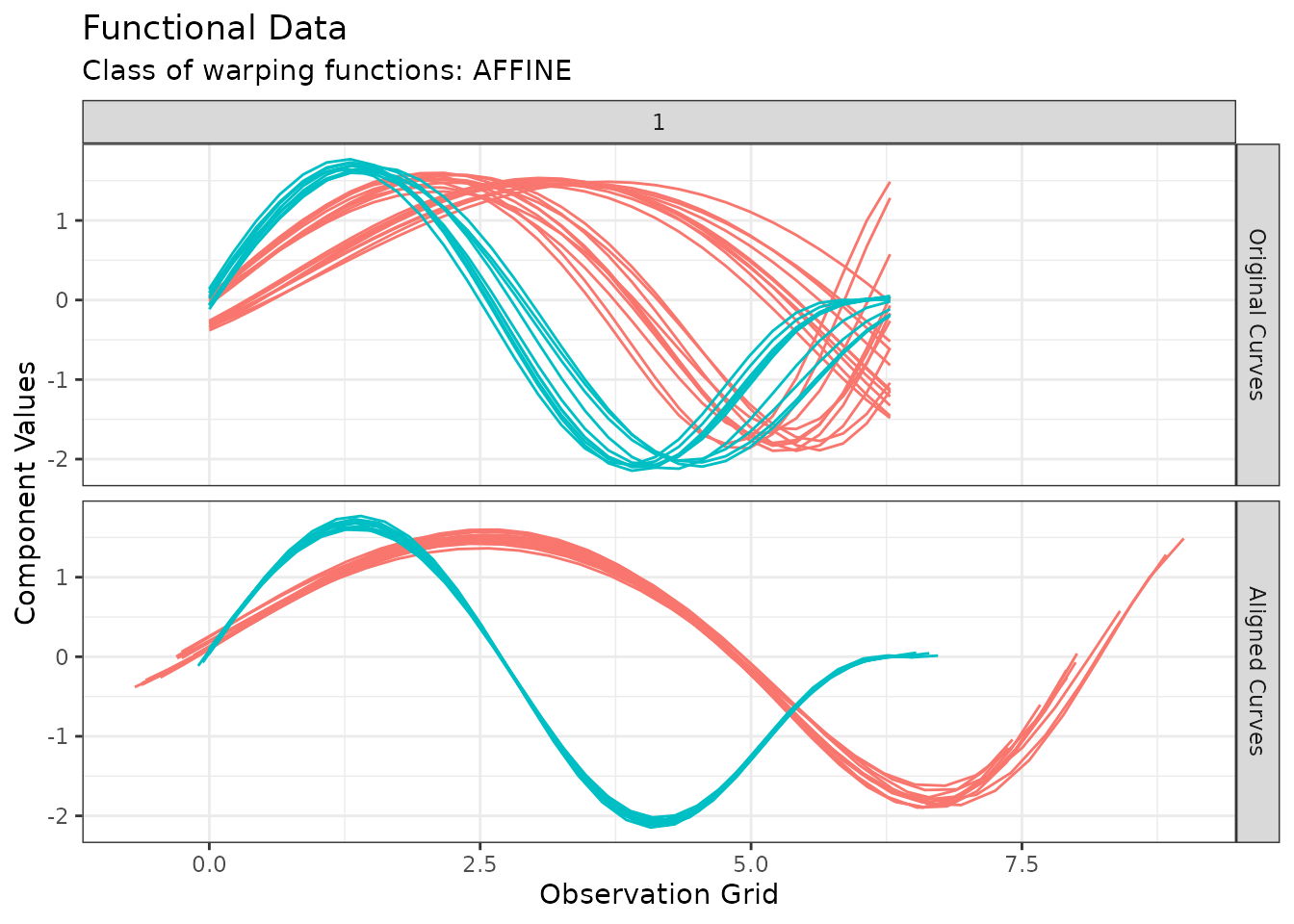
Or we can visualize the estimated warping functions with:
plot(out1, type = "phase")
#> Warning in data.frame(grid = as.vector(x$original_grids), value =
#> as.vector(x$aligned_grids), : row names were found from a short variable and
#> have been discarded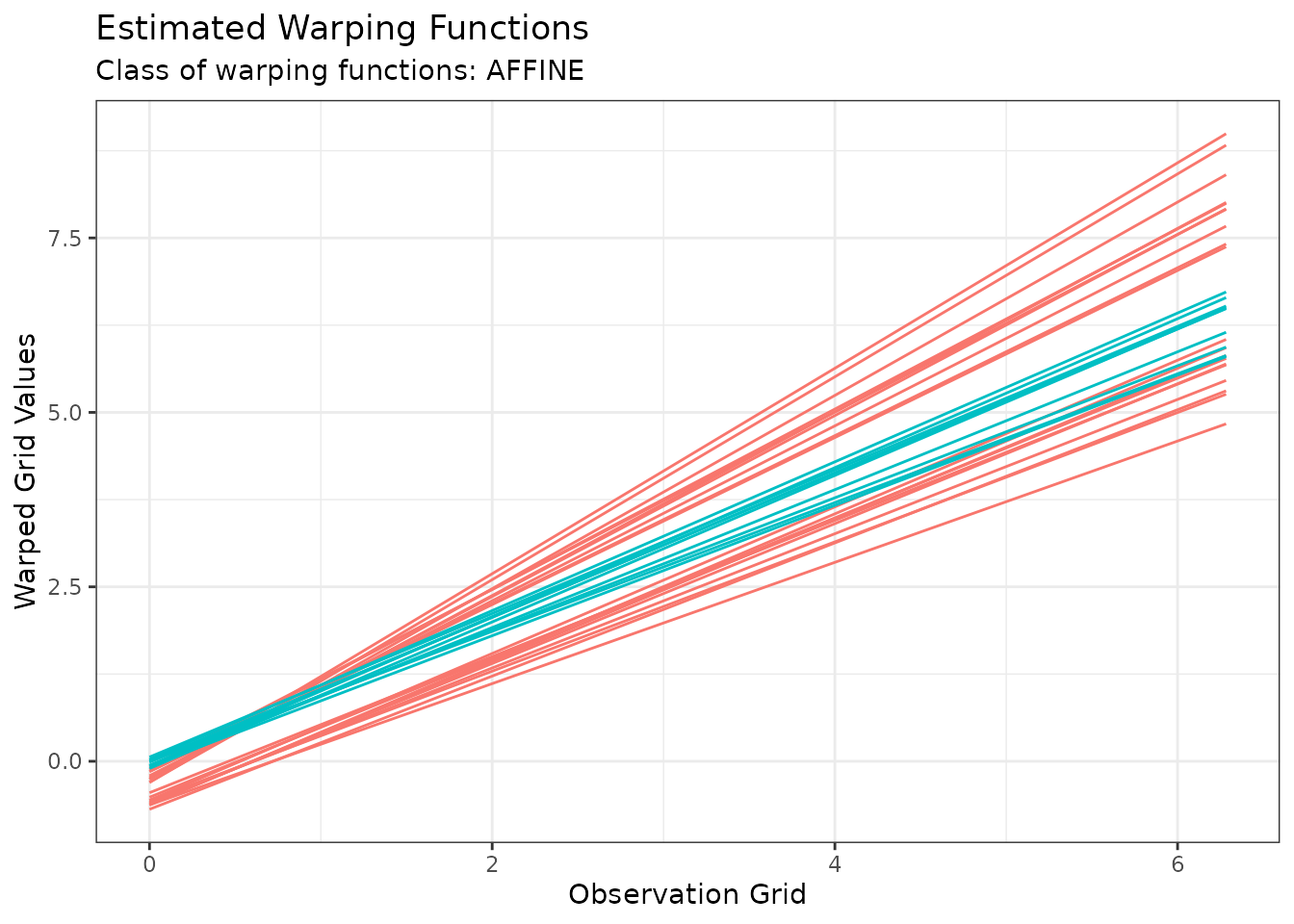
Or we can visualize the distribution of distances-to-center and silhouette values across observations with:
diagnostic_plot(out1)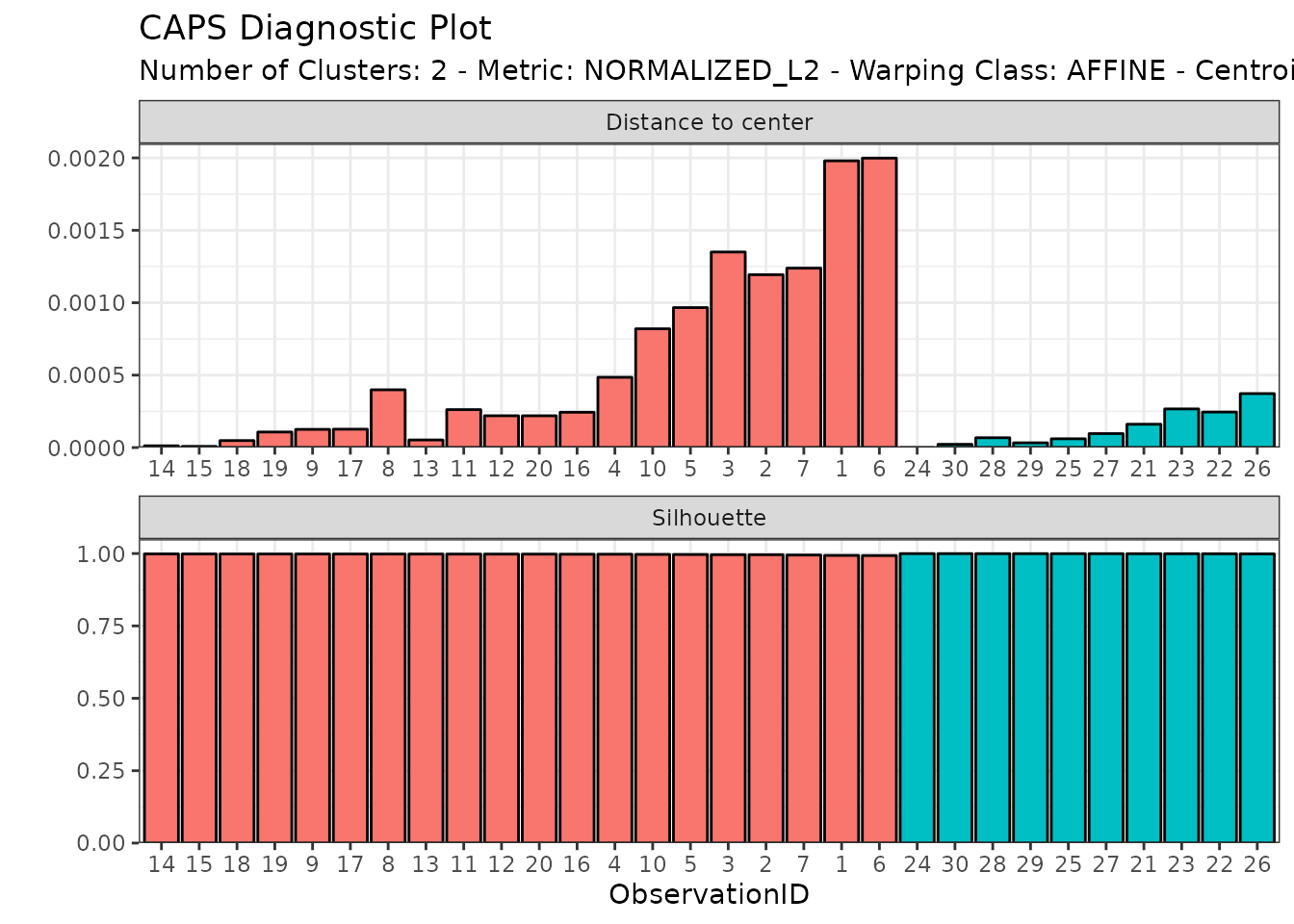
HAC based on phase variability
We can perform HAC based on phase variability only by switching the
cluster_on_phase argument to TRUE:
out2 <- fdahclust(
simulated30_sub$x,
simulated30_sub$y,
n_clusters = 3,
centroid_type = "mean",
warping_class = "affine",
metric = "normalized_l2",
cluster_on_phase = TRUE
)We can inspect the result as before:
plot(out2, type = "amplitude")
#> Warning in data.frame(grid = as.vector(grids), value = as.vector(unicurves), :
#> row names were found from a short variable and have been discarded
#> Warning in data.frame(grid = as.vector(grids), value = as.vector(unicurves), :
#> row names were found from a short variable and have been discarded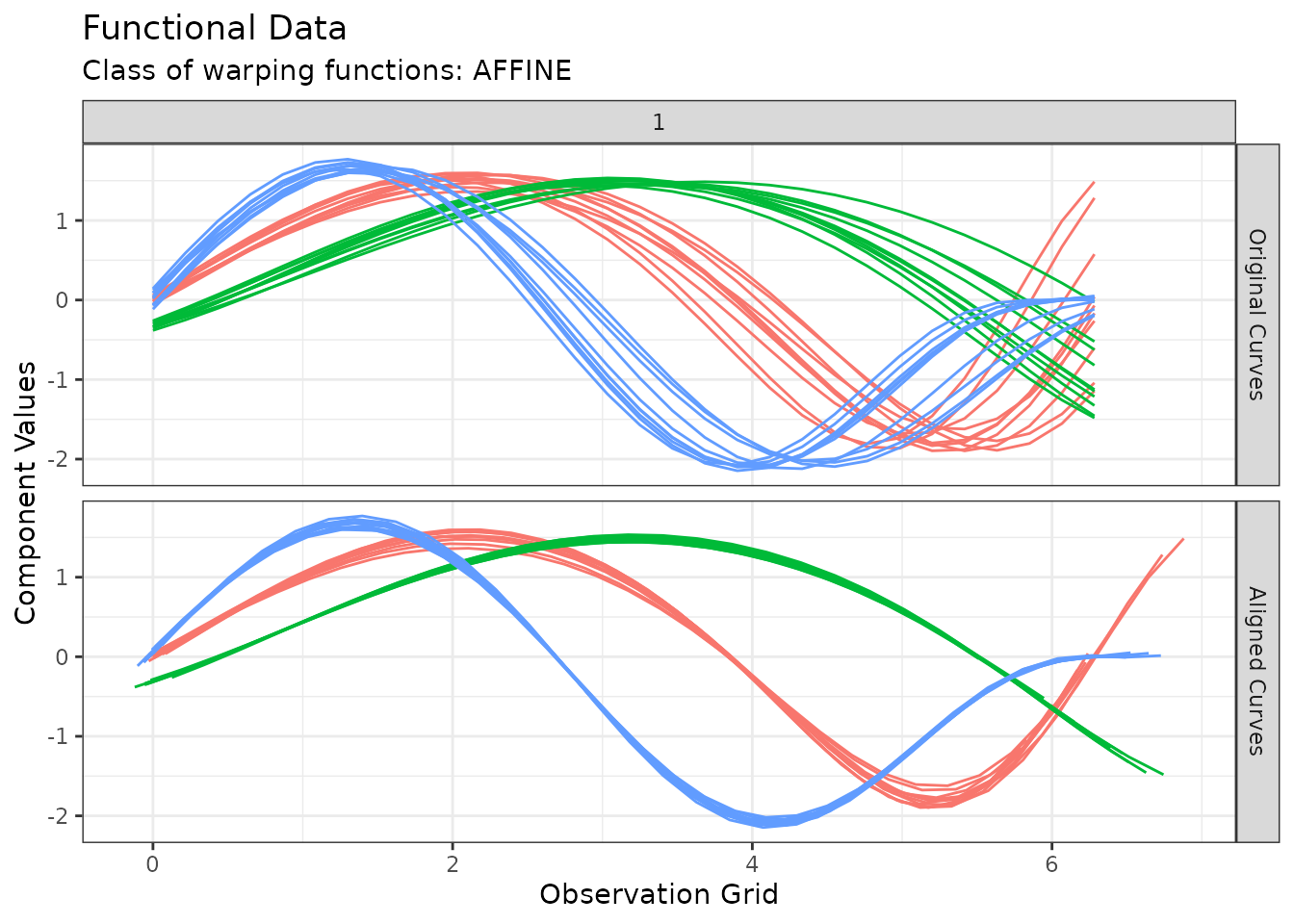
plot(out2, type = "phase")
#> Warning in data.frame(grid = as.vector(x$original_grids), value =
#> as.vector(x$aligned_grids), : row names were found from a short variable and
#> have been discarded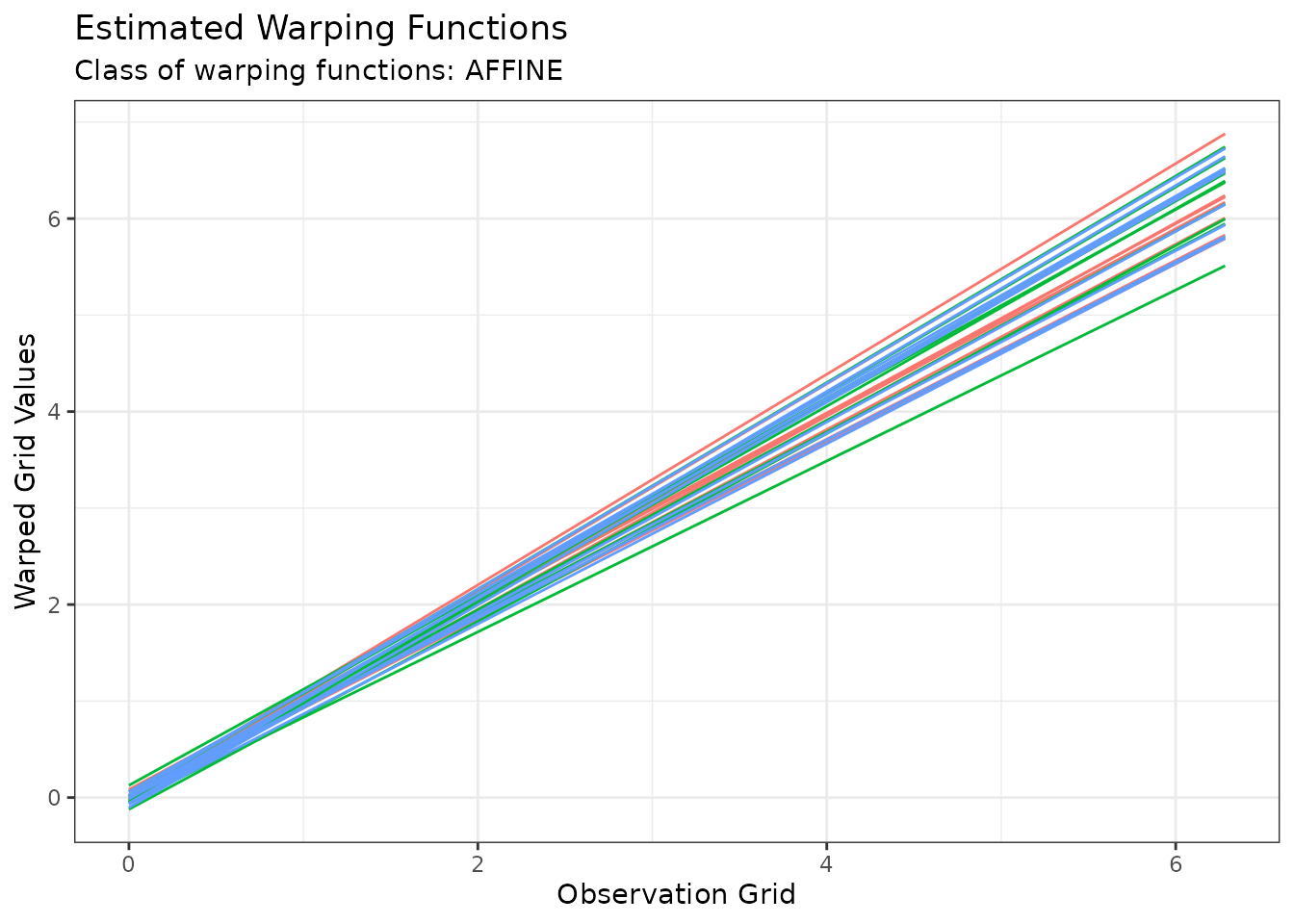
diagnostic_plot(out2)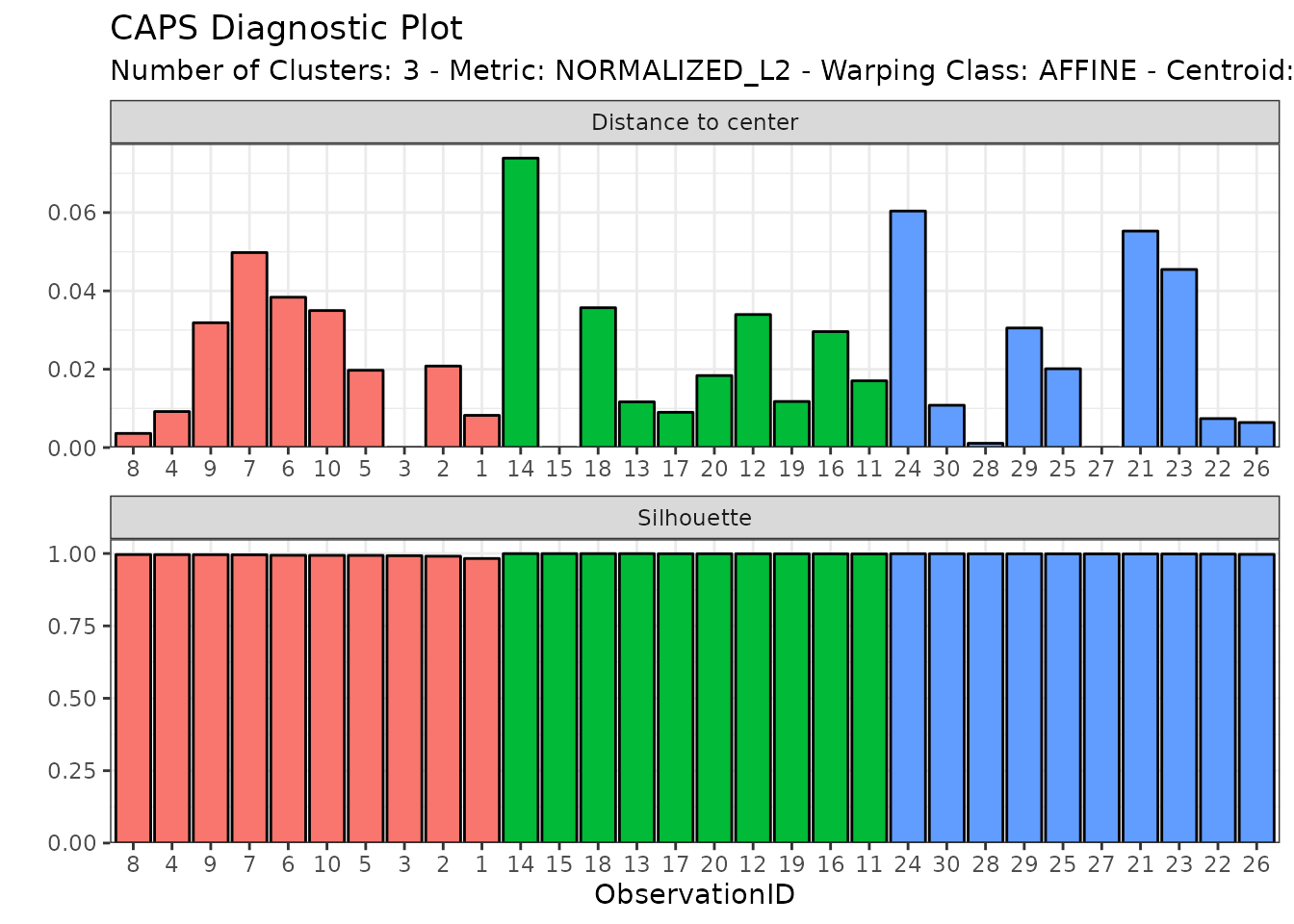
Choosing the number of clusters
The helper function compare_caps()
can be used to get an intuition of the number of clusters that one
should be looking for.
For example, we can generate data to help choosing the number of clusters when clustering on amplitude data:
ncores <- max(parallel::detectCores() - 1L, 1L)
plan(multisession, workers = ncores)
amplitude_data <- compare_caps(
x = simulated30_sub$x,
y = simulated30_sub$y,
n_clusters = 1:5,
metric = "normalized_l2",
warping_class = c("none", "shift", "dilation", "affine"),
clustering_method = "hclust-complete",
centroid_type = "mean",
cluster_on_phase = FALSE
)
plan(sequential)In this example. we asked through the specification of the optional
argument warping_class to use multiple warping classes and
store the clustering results separately in the output for later
comparison. Multiple choices can be also used for arguments
clustering_method and centroid_type although
in the above example we chose to focus on the HAC with complete linkage
using the mean as centroid type. The metric argument
must however be unique as we need to use the same
metric among methods for later comparison.
The above code generates an object of class mcaps which
has dedicated S3 specializations of
ggplot2::autoplot() and graphics::plot() as
well. These methods gain two extra optional arguments:
-
validation_criterion: A string specifying the validation criterion to be used for the comparison. Choices are"wss"or"silhouette". Defaults to"wss". -
what: A string specifying the kind of information to display about the validation criterion. Choices are"mean"(which plots the mean values) or"distribution"(which plots the boxplots). Defaults to"mean".
For example, one can can visualize the mean WSS for all specified methods as:
plot(amplitude_data, validation_criterion = "wss", what = "mean")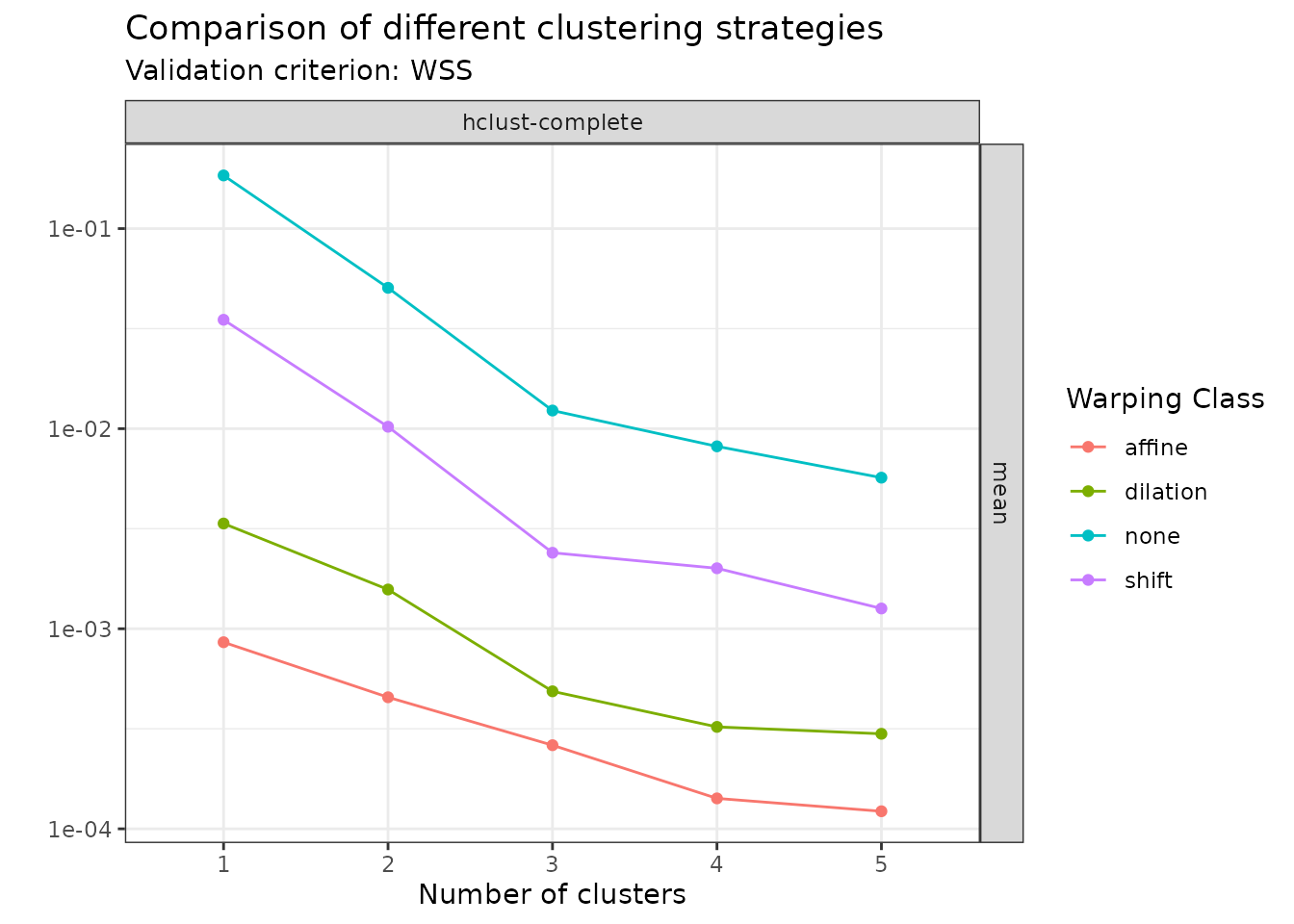
In this case, the plot clearly shows that among the set of warping classes that has been considered, one should use affine warping and search for groups when the goal is to cluster based on amplitude variability.
Let us now perform the same analysis with the goal of clustering based on phase variability instead:
ncores <- max(parallel::detectCores() - 1L, 1L)
plan(multisession, workers = ncores)
phase_data <- compare_caps(
x = simulated30_sub$x,
y = simulated30_sub$y,
n_clusters = 1:5,
metric = "normalized_l2",
warping_class = c("shift", "dilation", "affine"),
clustering_method = "hclust-complete",
centroid_type = "mean",
cluster_on_phase = TRUE
)
plan(sequential)
plot(phase_data, validation_criterion = "wss", what = "mean")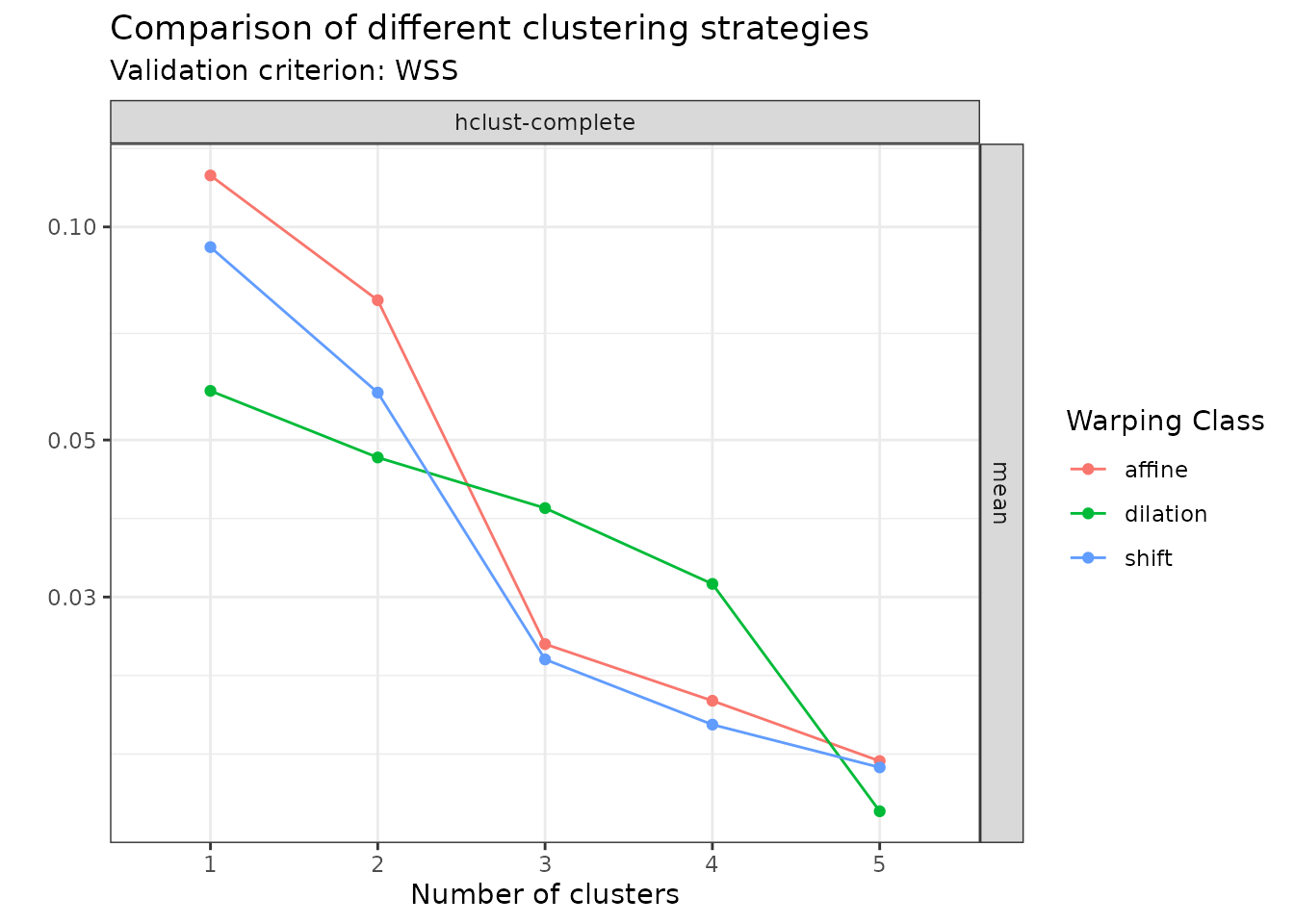
In this case, as expected by looking at the original data set, the plot suggests to search for groups. What is even more interesting is that there is in this case no improvement in terms of WSS by going from the shift warping class to the affine warping class. This was also expected by looking at the original data set but it is nice that it is also reflected in the output diagnostic.