fdacluster
An R package for joint clustering and alignment of functional data
Department of Mathematics Jean Leray, UMR CNRS 6629, Nantes University, France
MOX - Department of Mathematics, Politecnico di Milano, Italy
2023-06-22
Title explanation
Clustering
- Unlabeled data: observations are not assigned to pre-existing groups;
- Goal: group together individuals or observations that share common features while ensuring the following property:
- observations within a group must be as similar as possible,
- observations between groups (belonging to different groups) must be as different as possible.
Clustering methods
Centroid-based clustering
Clusters are represented by a central value or centroid. This centroid might not necessarily be one of the observations. These methods (e.g. k-means) operate in an iterative fashion in which the notion of similarity is derived by how close an observation is to the centroid of the cluster.
Hierarchical clustering
Observations that are closer in the data space are more related (similar) than observations that are far from each other. Clusters are formed by connecting observations according to their distance and clusters are allowed to be merged or splitted according to a cluster-based distance.
Density-based clustering
Search the data space for areas of high density of observations (e.g. DBSCAN). Clusters are defined as the areas of higher density within the data space compared to other regions. Observations in the sparse areas are usually considered to be noise and/or border points.
A common ingredient
All methods but model-based rely on the ability to quantify closeness / distance between observations.
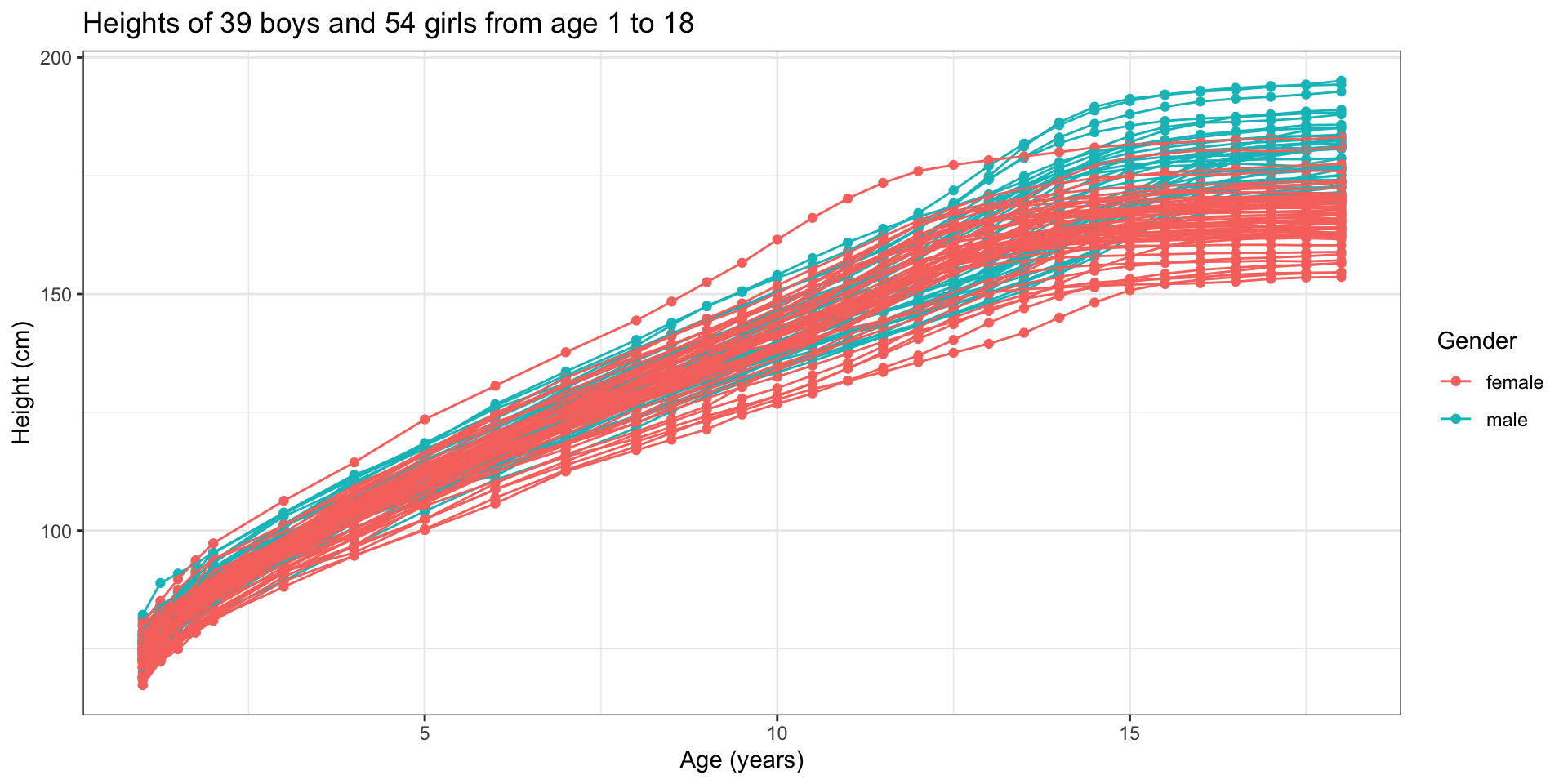
Functional data - Visual definition
Data source: Berkeley Growth Study data (Tuddenham and Snyder 1954; Ramsay and Silverman 2006)
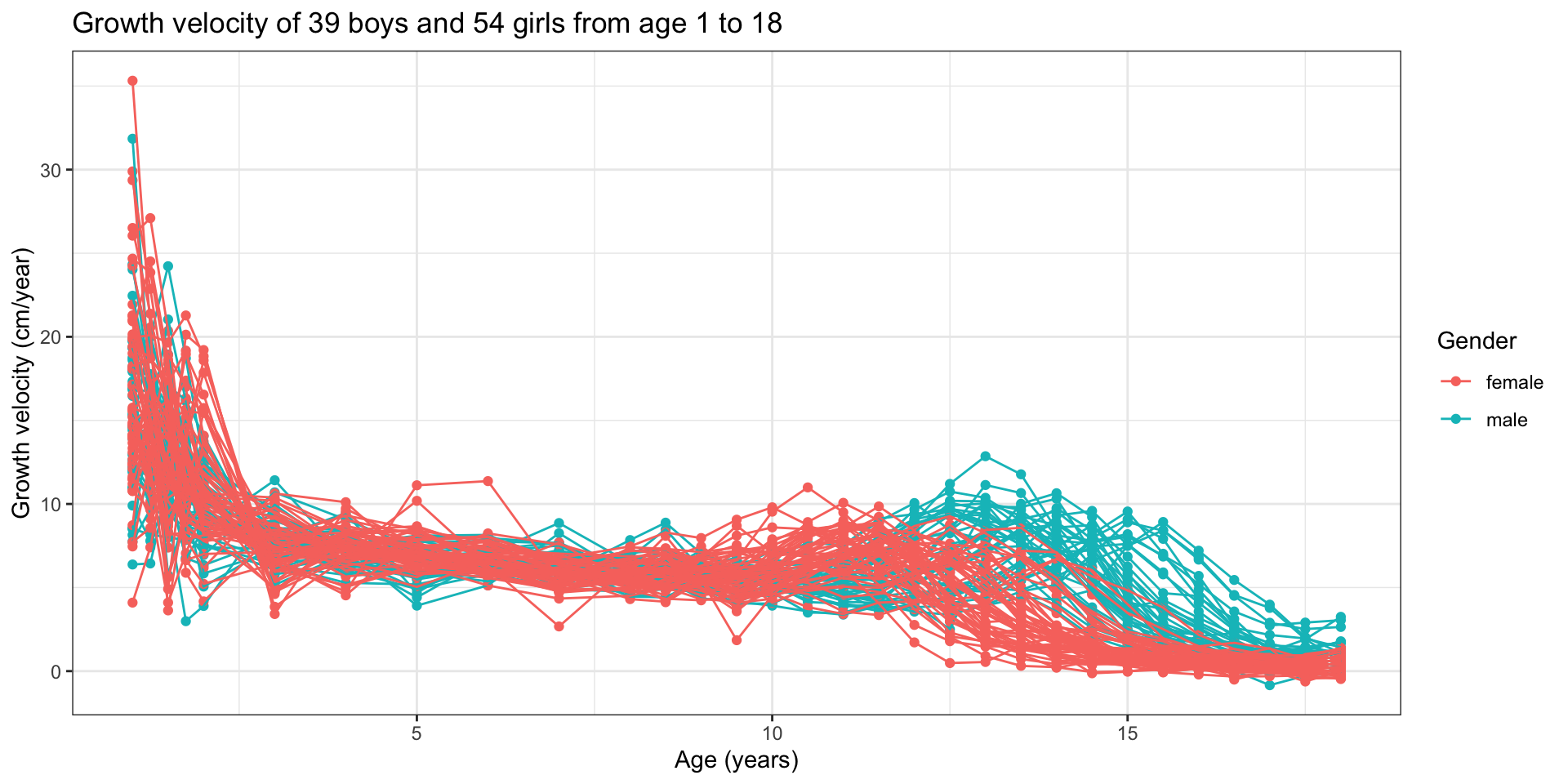
Functional data - Derivatives
What if we did not know gender membership?
- Common peak of growth velocity shortly after birth
- Another smaller peak at puberty occurring:
- At different ages for girls and for boys;
- At different ages for each individual.
Functional clustering
Functional data - Notations
- We consider a random variable \(X\) which takes values in \(F = \{ f: \mathcal{D} \to \mathcal{M} \}\).
- The domain \(\mathcal{D}\) of the functions in \(F\) is a subset of \(\mathbb{R}\). Typically \(\mathcal{D} = \mathbb{R}\) or \(\mathcal{D} = [a, b]\).
- The co-domain \(\mathcal{M}\) of the functions in \(F\) can be univariate or multivariate and can be either a Euclidean space or more generally a manifold. Typically, \(\mathcal{M} = \mathbb{R}^p\).
- The space \(F\) is equipped with a metric \(d\) so that \((F, d)\) is a metric space.
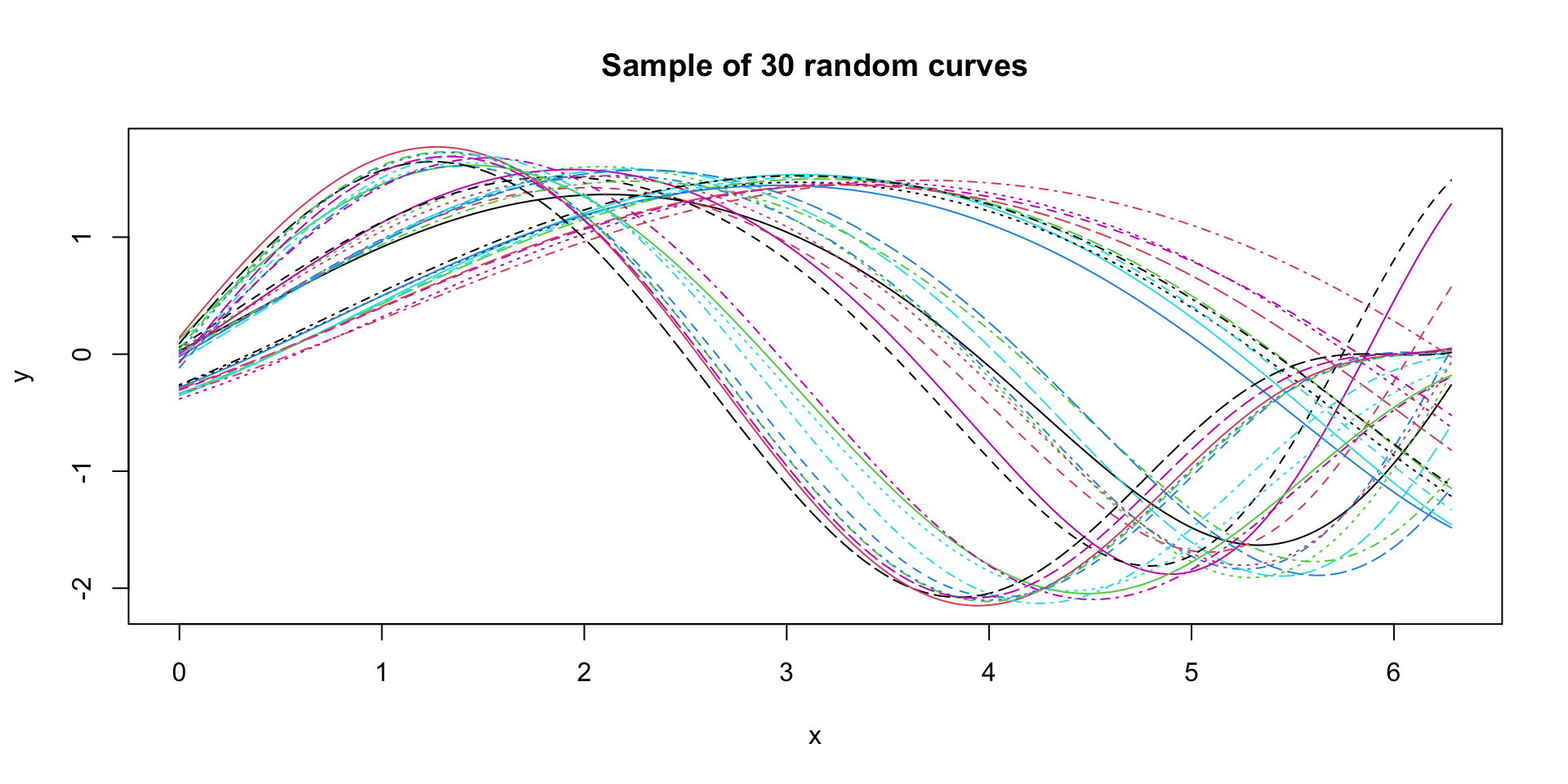
Functional data - A simpler example
Data source: Sangalli et al. (2010).
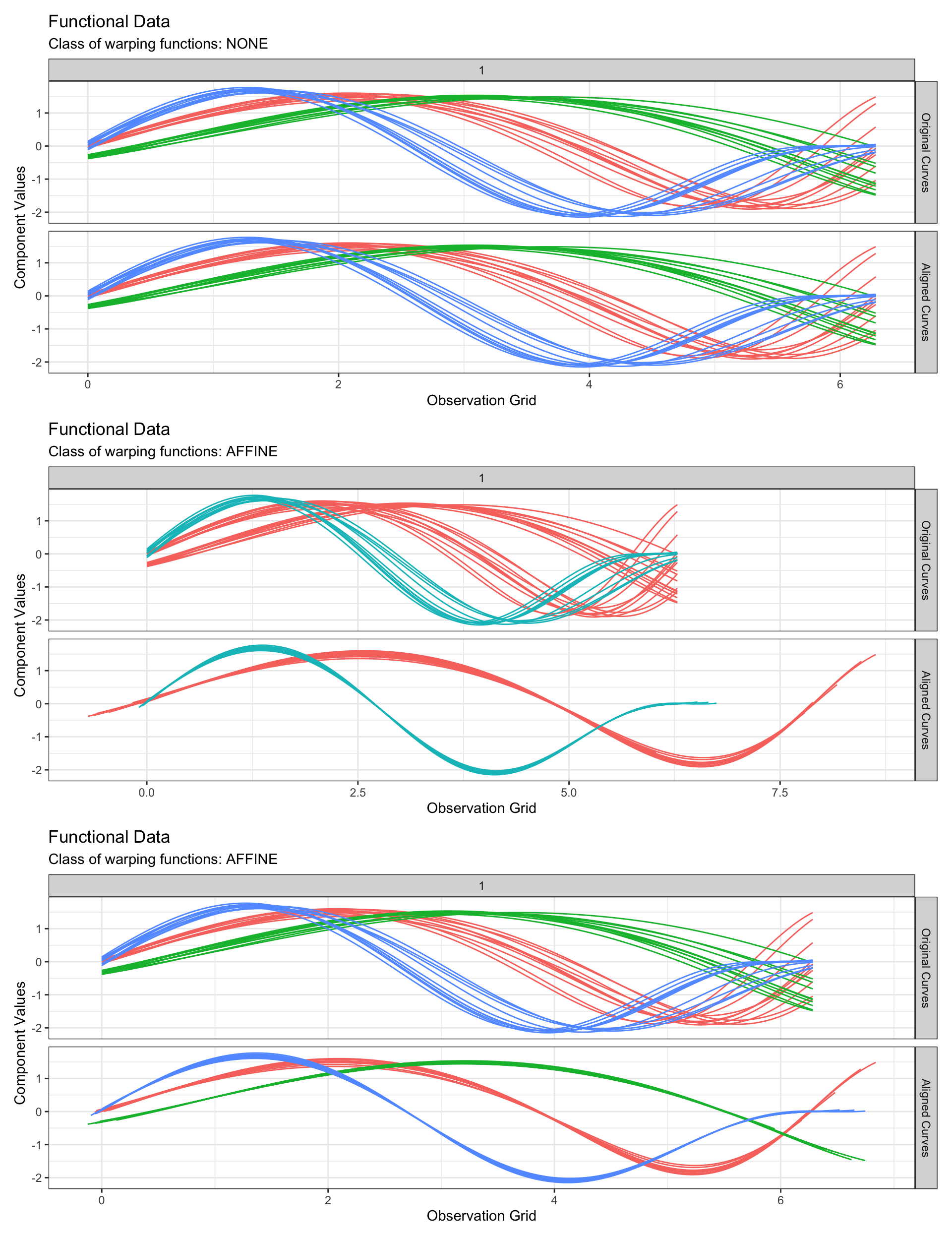
Functional data - Clustering
Solution 1: \[ \begin{cases} d_A^2(f, g) = d^2(f, g) \\ d_C^2(f, g) = d_A^2(f, g) \end{cases} \]
Solutions 2 & 3: \[ \begin{cases} d_A^2(f, g) = \min_{h \in W} d^2(f \circ h, g) \\ d_C^2(f, g) = d_A^2(f, g) \end{cases} \]
\[ \begin{cases} d_A^2(f, g) = \min_{h \in W} d^2(f \circ h, g) \\ d_C^2(f, g) = \int_{\mathcal{D}} \left[ h^\star(t) - t \right]^2 dt \\ \end{cases} \] with \(h^\star(t) = \arg \min_{h \in W} d^2(f \circ h, g)\).
Registration of curves
The following properties on the functional space \(F\), its metric \(d\) and the set \(W\) of warping functions considered for curve registration must be satisfied for a mathematically consistent registration setup (Vantini 2012):
\(F = \{ f: \mathcal{D} \to \mathcal{M} \}\) is a metric space equipped with a metric \(d: F \times F \to \mathbb{R}^+\);
\(W\) is a subgroup — with respect to ordinary composition \(\circ\) — of the group of the continuous automorphisms of \(\mathcal{D}\);
\(\forall f \in F\) and \(\forall h \in W\), we have that \(f \circ h \in F\);
Given any couple of elements \(f_1, f_2 \in F\) and an element \(h \in W\), the distance between \(f_1\) and \(f_2\) is invariant under the composition of \(f_1\) and \(f_2\) with \(h\), i.e.:
\[ d(f_1, f_2) = d(f_1 \circ h, f_2 \circ h). \]
We will refer to this property as \(W\)-invariance of \(d\).
The case \(\mathcal{D} = \mathbb{R}\)
In this situation, it is not possible to perform registration over \(\mathbb{R}\) and most of the time we use \(W\) to be the set of affine warping functions i.e. \(W = \{ h: h(t) = at+b \}\).
In this setting, it is tempting to equip \(F\) with the \(L^2\)-distance: \[ d_{L^2}^2(f, g) = \int_\mathbb{R} \left\| f(t) - g(t) \right\|_{\mathbb{R}^p}^2 dt. \]
The \(L^2\)-distance is however not affine-invariant:
\[ d_{L^2}^2(f \circ h, g \circ h) = \frac{1}{a} d_{L^2}^2(f, g) \]
A proper normalization can make it affine-invariant such as: \[ d(f, g) = \frac{d_{L^2}(f, g)}{\| f \|_{L^2} + \| g \|_{L^2}} \]
The case \(\mathcal{D} = [a, b]\)
In this situation, it is possible to perform registration over the set \(W\) of all boundary-preserving warping functions by using the square-root slope function (SRSF) framework (Tucker, Wu, and Srivastava 2013) under the assumption that all functions \(f \in F\) are absolutely continuous. In this case, the SRSF of a function \(f \in F\) which reads:
\[ \begin{array}{rccl} q_f : & \mathbb{R} & \to & \mathbb{R}^p \\ & t & \mapsto & \frac{\dot{f}(t)}{\sqrt{ \| \dot{f}(t) \|_{\mathbb{R}^p} }}, \end{array} \] is square-integrable and thus belong to \(L^2([a,b], \mathbb{R}^p)\).
We can therefore define: \[ d(f, g) = d_{L^2}(q_f, q_g), \] and one can show that \(d\) is invariant under any boundary-preserving warping.
Funct. \(k\)-means (Sangalli et al. 2010)
Initialization
Choose \(k\) initial observations as cluster centroids.
Align observations
Perform registration of all observed curves on each cluster centroid.
Assign observations to closest centroid
Assign each observed curve to the cluster with smallest distance to its center. This involves solving a constrained mixed integer and linear programming problem to ensure that no requested cluster will be empty.
Update cluster centroids
Recompute each cluster centroid given the newly assigned observations. Since the solution to the registration problem is not unique, the unique centroid is computed according to recommendations in Vantini (2012) to minimize total variation. If \(f_0\) is a computed centroid, then define the cluster centroid as \[ \widehat{f_0} = \arg \min_{f \in [f_0]} \sum_{i=1}^n d^2(f_i, f), \] which is achieved for some warping function \(h^\star\) which needs to be further applied to all observations in that cluster.
Funct. hierarchical clustering
Distance matrix computation
Compute the matrix \(D\) of pairwise distances between curves minimizing over the selected class \(W\) of warping functions.
Dendrogram computation
Apply traditional approaches to hierarchical clustering on \(D\) (either agglomerative or divisive) to build a hierarchy of possible clustering partitions of the data (dendrogram).
Assign observations to clusters
- Cut the dendrogram to form \(k\) clusters and, in each cluster;
- Register all curves to centroid via funct. \(1\)-mean clustering.
Update distance matrix
Update the distance matrix \(D\) using the registered curves from previous step with no further registration. This is used for final calculations of WSS and silhouettes.
Functional DBSCAN
Adapting DBSCAN to functional data follows the same principles.
The R package fdacluster
On CRAN (or almost)
You can install the official version from CRAN via:
or you can opt to install the development version from GitHub with:
Test it out now!
If you want to test the package right now, use the development version.
Features
A dedicated
S3class for storing results of clustering with amplitude and phase separation;Implementations of k-means, HAC and DBSCAN methods for functional data;
Built-in parallelization;
Tools for comparing various clustering strategies:
compare_caps(): generates an object of classmcaps()containingcapsobjects for several clustering strategies;plot()andautoplot()S3 specialized methods formcapsobjects.
Integration within the R fda landscape: input grids and curves can be
Built-in parallelization
- Achieved via the futureverse framework;
- Possibility of displaying a progress bar.
library(future)
library(progressr)
handlers("rstudio") # Defines RStudio progress bar style
plan(multisession, workers = parallelly::availableCores(omit = 1)) # Turns ON parallelization
with_progress(
# User-defined code for clustering
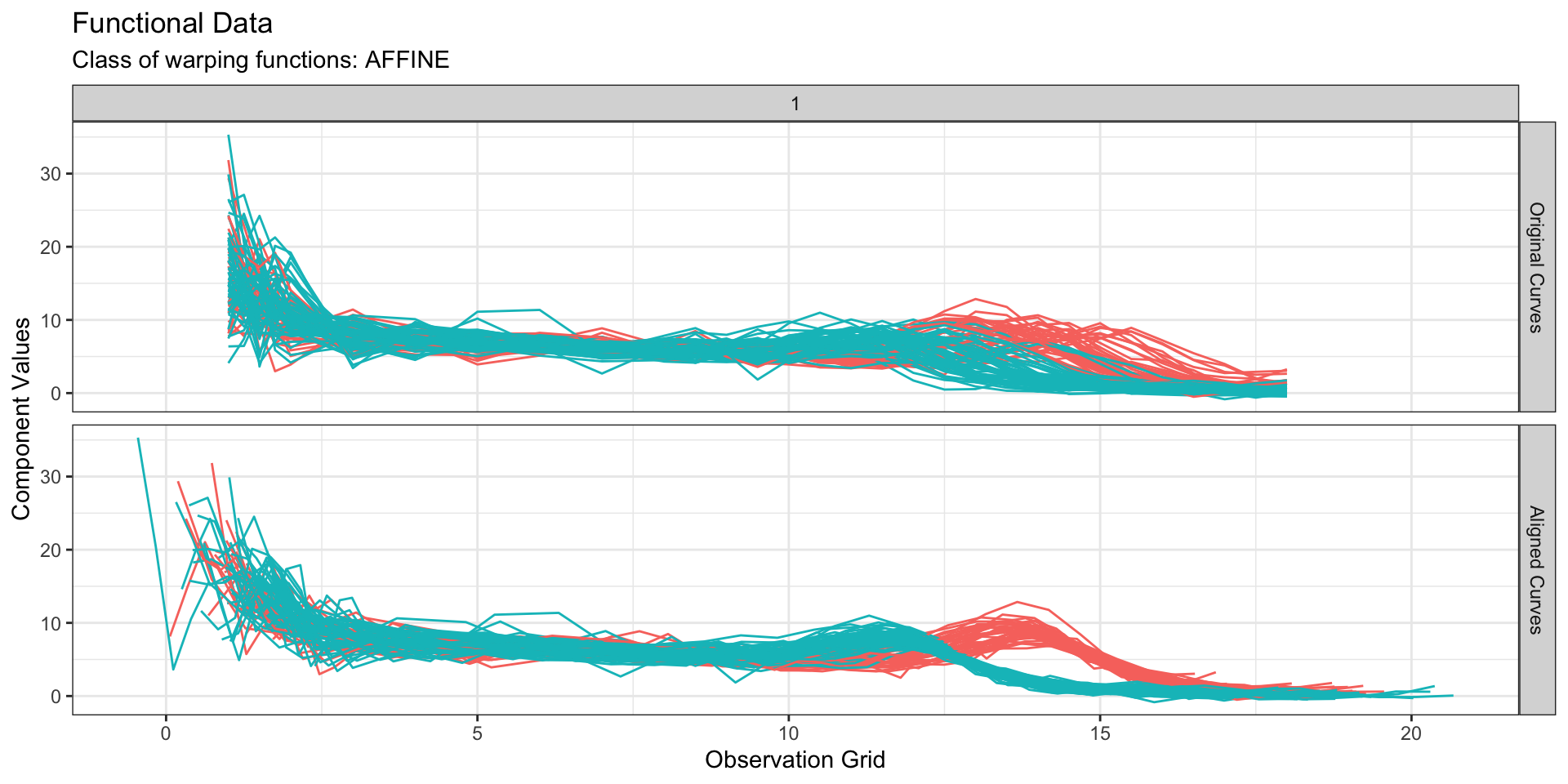
out1 <- fdahclust(
x = x,
y = t(y1),
n_clusters = 2,
metric = "normalized_l2",
warping_class = "affine",
cluster_on_phase = TRUE
)
)
plan(sequential) # Turns OFF parallelizationThe caps class
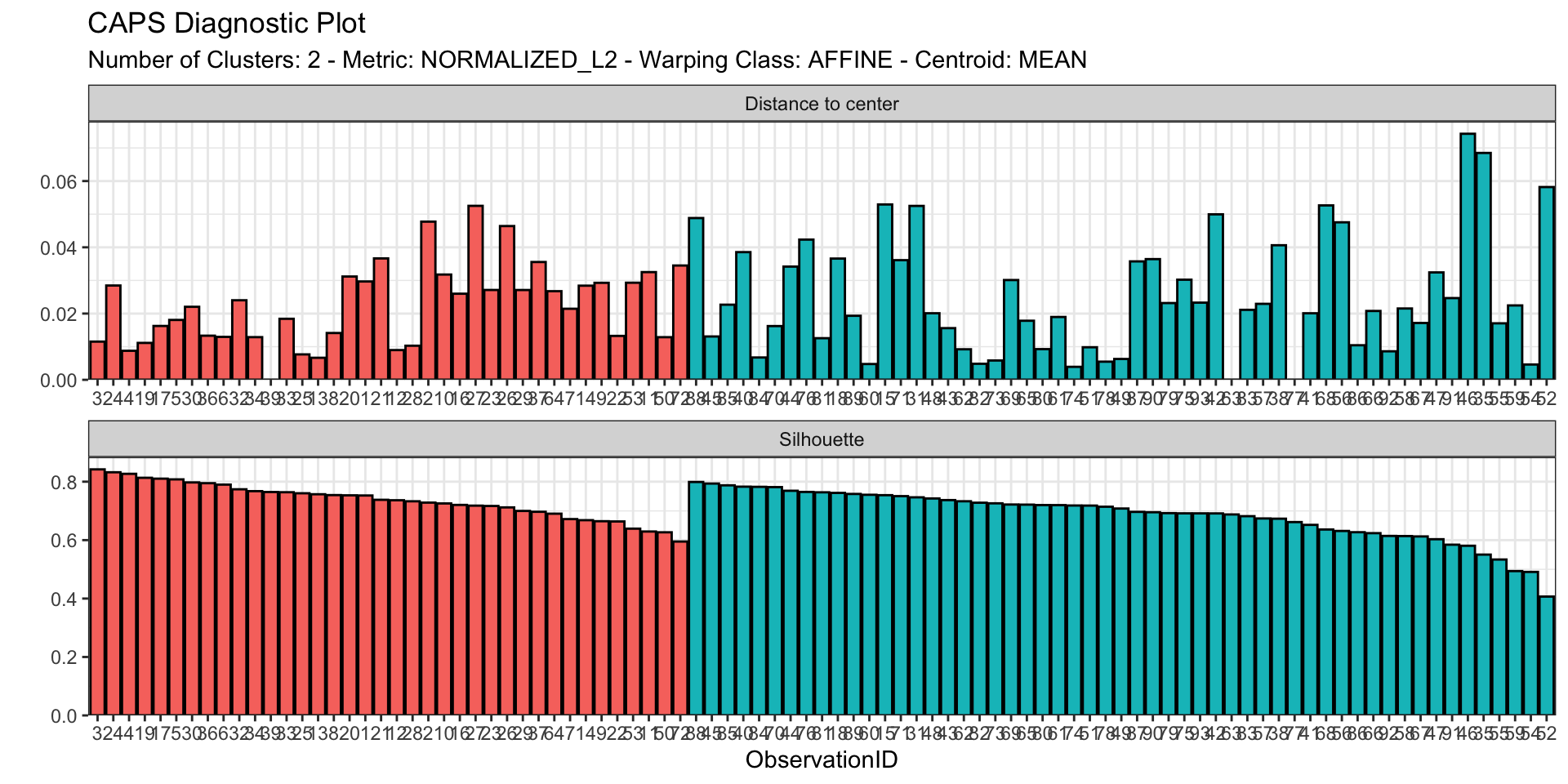
List of 14
$ original_curves : num [1:93, 1, 1:31] 12.44 18.13 12.02 16.92 8.51 ...
$ original_grids : num [1:93, 1:31] 1 1 1 1 1 1 1 1 1 1 ...
$ aligned_grids : num [1:93, 1:31] 1.527 1.275 1.845 0.937 2.262 ...
$ center_curves : num [1:2, 1, 1:31] 20.3 17.6 16.5 15.3 12.6 ...
$ center_grids : num [1:2, 1:31] 0.26 0.307 0.898 0.968 1.537 ...
$ n_clusters : num 2
$ memberships : Named int [1:93] 1 1 1 1 1 1 1 1 1 1 ...
..- attr(*, "names")= chr [1:93] "1" "2" "3" "4" ...
$ distances_to_center: num [1:93] 0.02969 0.04775 0.01153 0.00876 0.01807 ...
$ silhouettes : num [1:93] 0.756 0.744 0.841 0.832 0.808 ...
$ amplitude_variation: num 0.0606
$ total_variation : num 0.141
$ n_iterations : num 0
$ call_name : chr "fdahclust"
$ call_args :List of 20
..$ x : symbol x
..$ y : language t(y1)
..$ n_clusters : num 2
..$ is_domain_interval : logi FALSE
..$ transformation : chr "identity"
..$ warping_class : chr "affine"
..$ centroid_type : chr "mean"
..$ metric : chr "normalized_l2"
..$ cluster_on_phase : logi TRUE
..$ linkage_criterion : chr "complete"
..$ use_verbose : logi FALSE
..$ warping_options : language c(0.15, 0.15)
..$ maximum_number_of_iterations: int 100
..$ number_of_threads : int 1
..$ parallel_method : int 0
..$ distance_relative_tolerance : num 0.001
..$ use_fence : logi FALSE
..$ check_total_dissimilarity : logi TRUE
..$ compute_overall_center : logi FALSE
..$ centroid_extra : num 0
- attr(*, "class")= chr [1:2] "caps" "list"Common interface for clustering functions
What about boys and girls?
| Cluster | female | male |
|---|---|---|
| 1 | 8.33% (3) | 91.67% (33) |
| 2 | 89.47% (51) | 10.53% (6) |
References

Rencontres R 2023 – Avignon – https://astamm.github.io – aymeric.stamm@cnrs.fr