midi
A Shiny app for designing diffusion MRI protocols
2024-06-14
Context
Brain microstructure
What is of interest ?
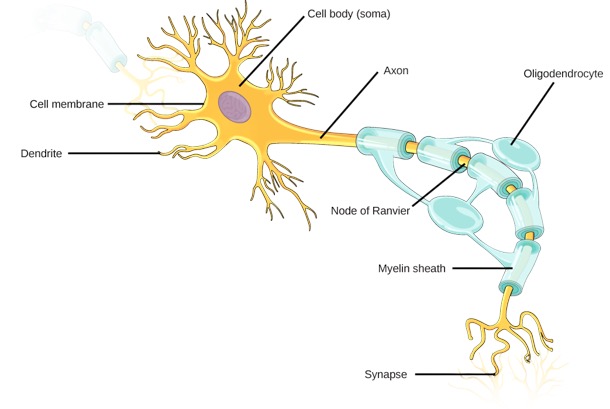
Neural transmission
Parameters of interest
- Axonal density: \(0.5-0.8\);
- Axon diameter: \(0.5-10\,\mu m\);
- Glial cell density: \(0.1-0.5\);
- Glial cell diameter: \(20-30\,\mu m\).
What can we see to the naked eye ?
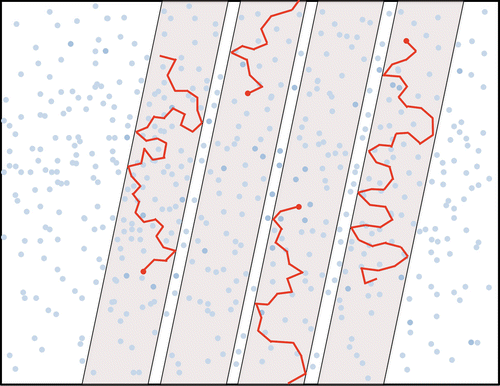
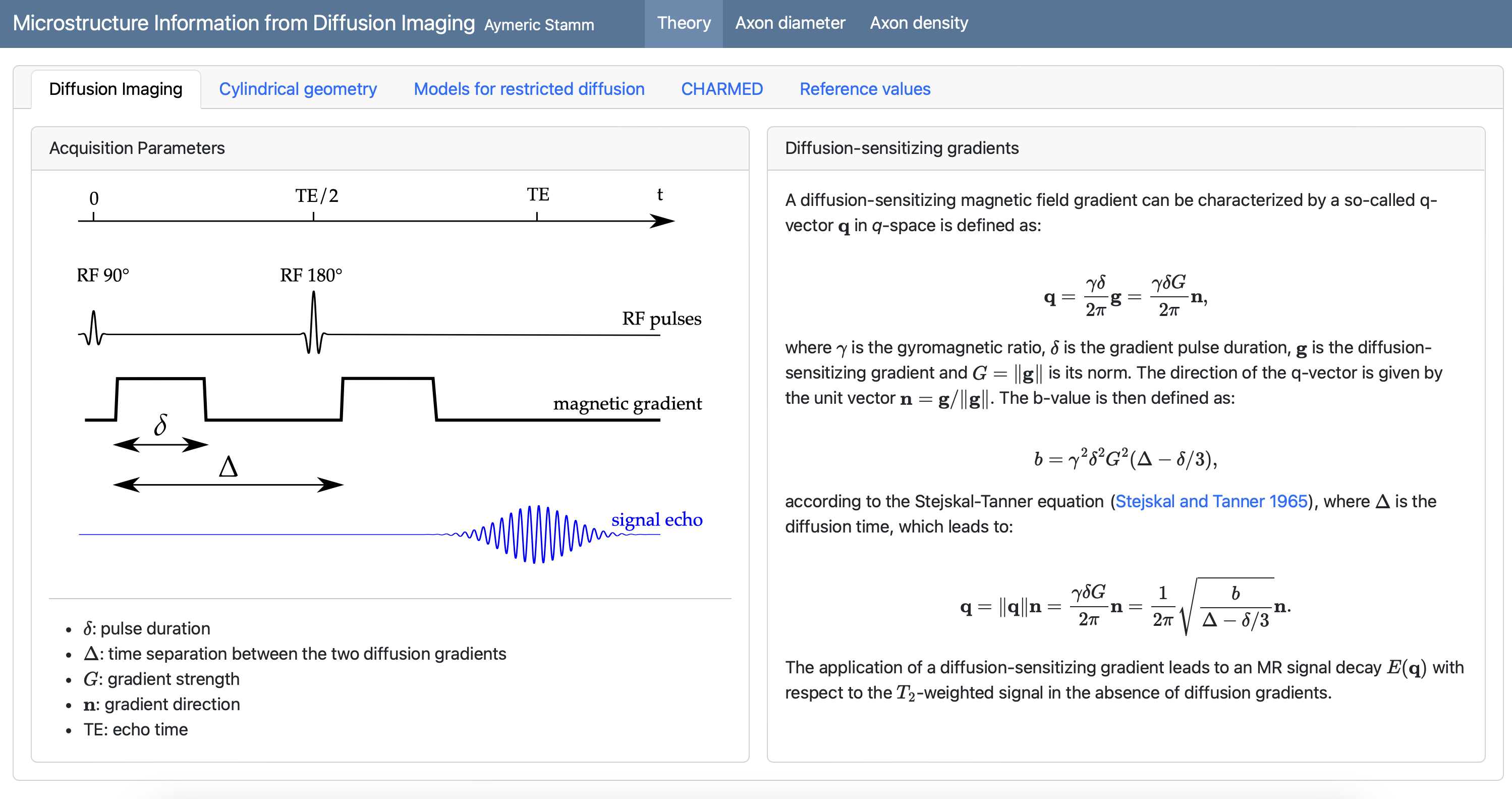
Principle of diffusion MRI
- The Human body is made of \(70\%\) of water.
- Water molecules are in constant motion due to thermal agitation.
- Water trapped in biological tissues is hindered by the presence of cell membranes, axons, dendrites, etc.
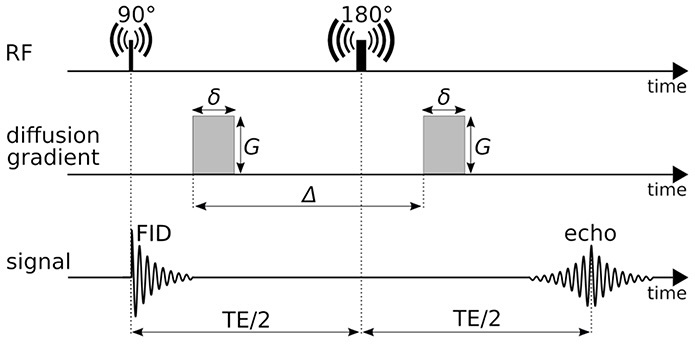
Principle of diffusion MRI (2/2)
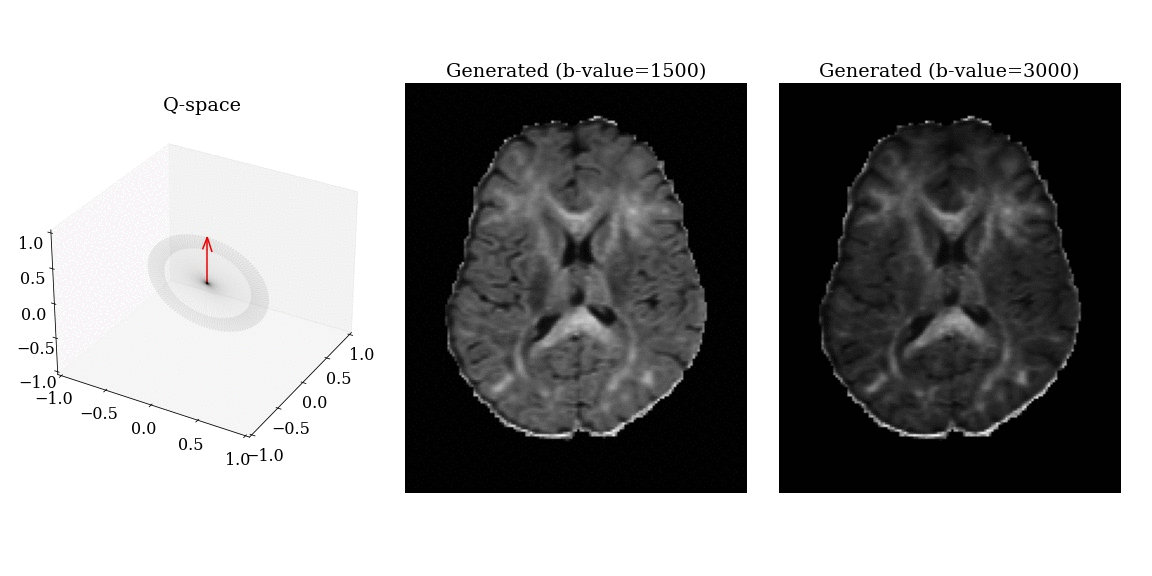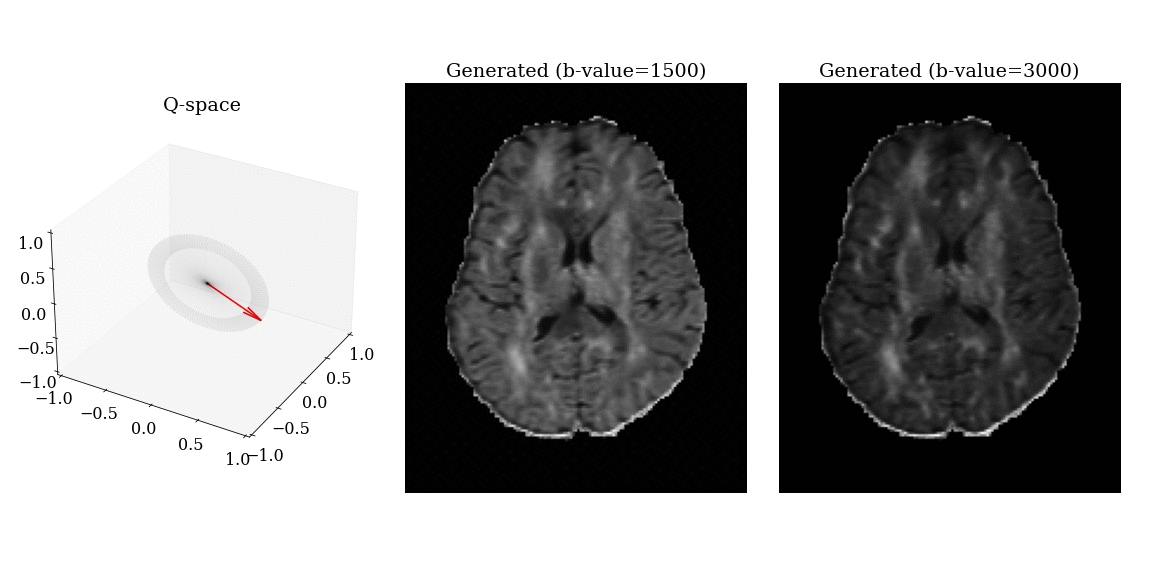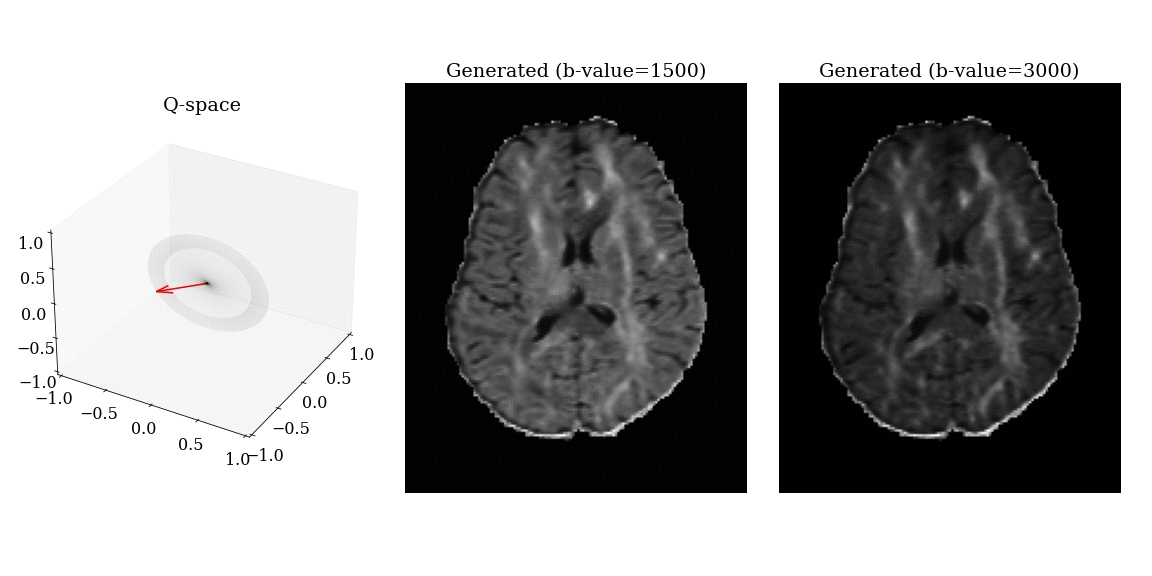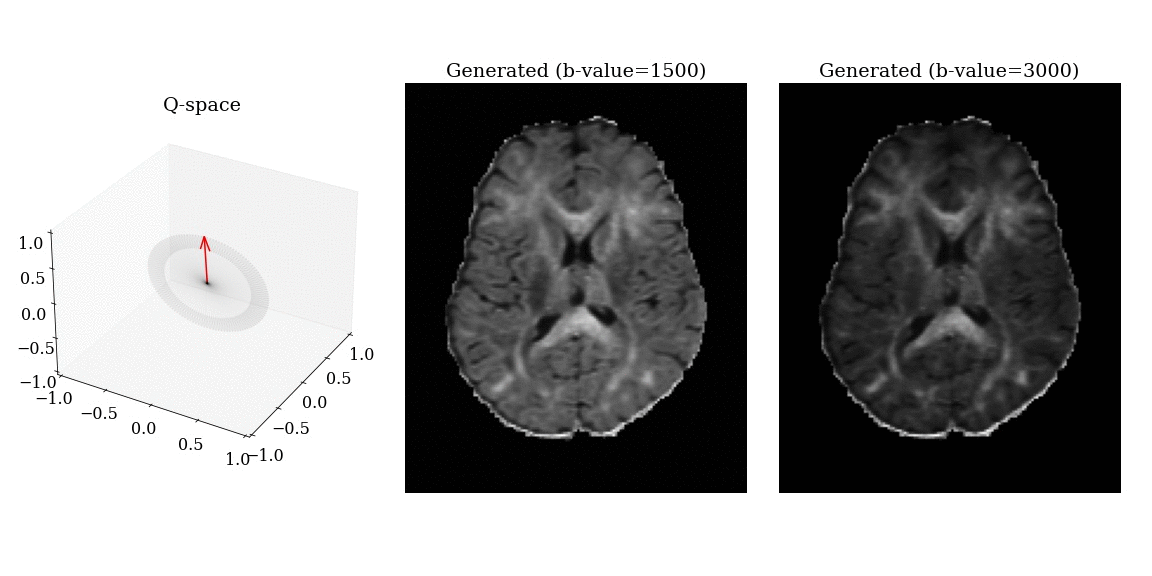
- Apply magnetic field gradients \(\mathbf{q}\) in different directions;
- Measure the signal attenuation \(S(\mathbf{q})\) induced by restricted diffusion;
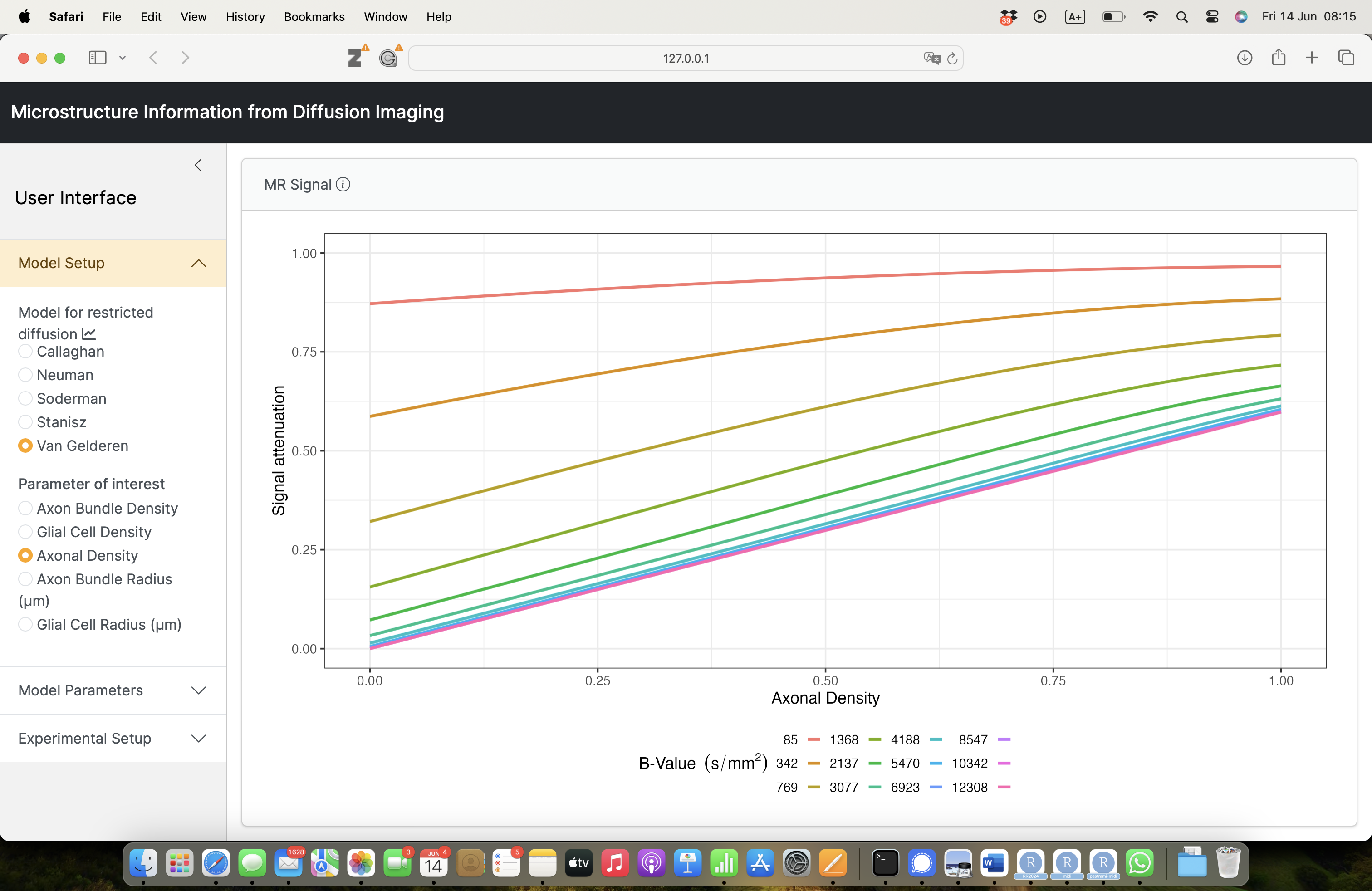
- Measured signal carries information on the microstructure.
\[ S(\mathbf{q}) = \int_{\mathbb{R}^3} \rho(\mathbf{r}) e^{-i \mathbf{q} \cdot \mathbf{r}} \, d\mathbf{r} \]
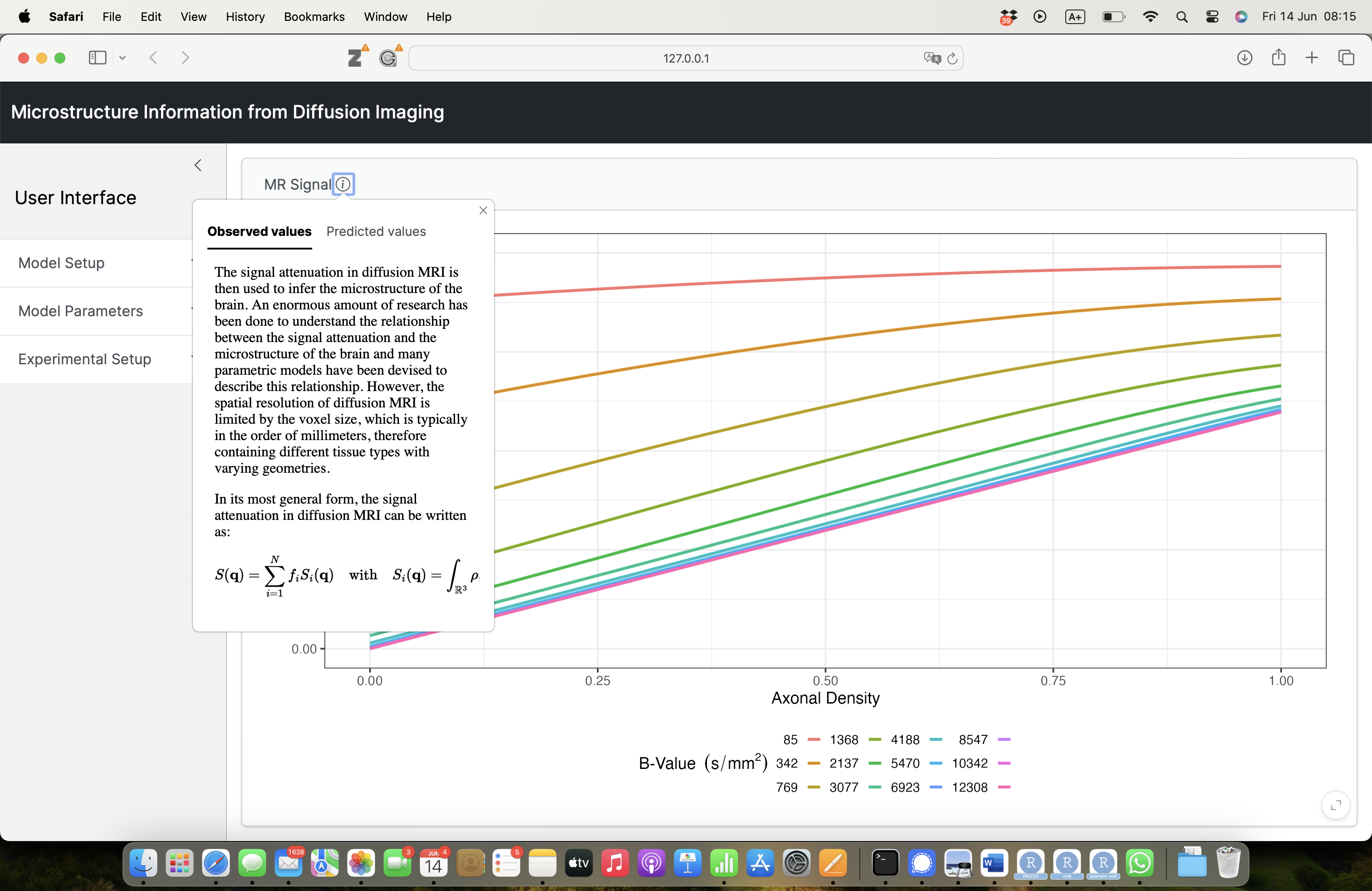
Voxel size VS microstructure
\[ S(\mathbf{q}) = \sum_{i=1}^N f_i S_i(\mathbf{q}) \quad \text{with} \quad S_i(\mathbf{q}) = \int_{\mathbb{R}^3} \rho_i(\mathbf{r}) e^{-i \mathbf{q} \cdot \mathbf{r}} \, d\mathbf{r} \]
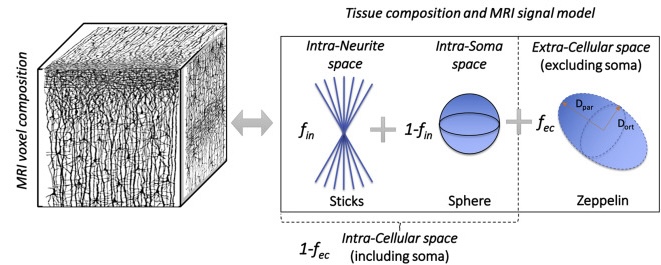
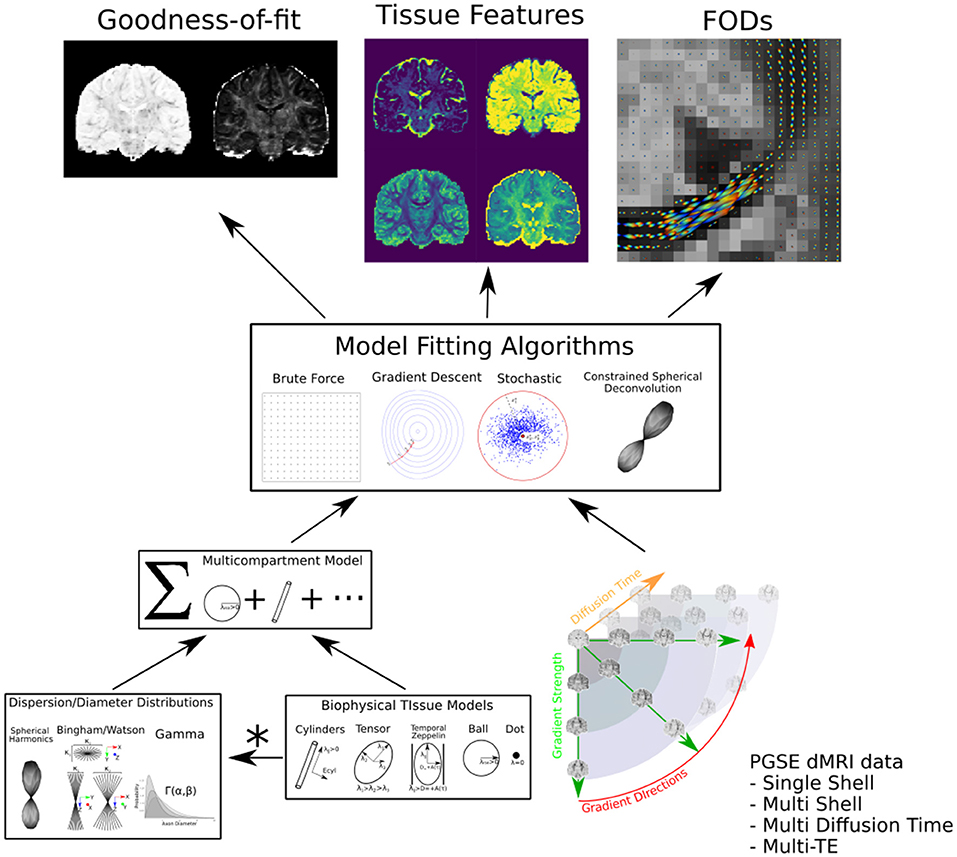
Compartment modeling
Data acquisition and modeling
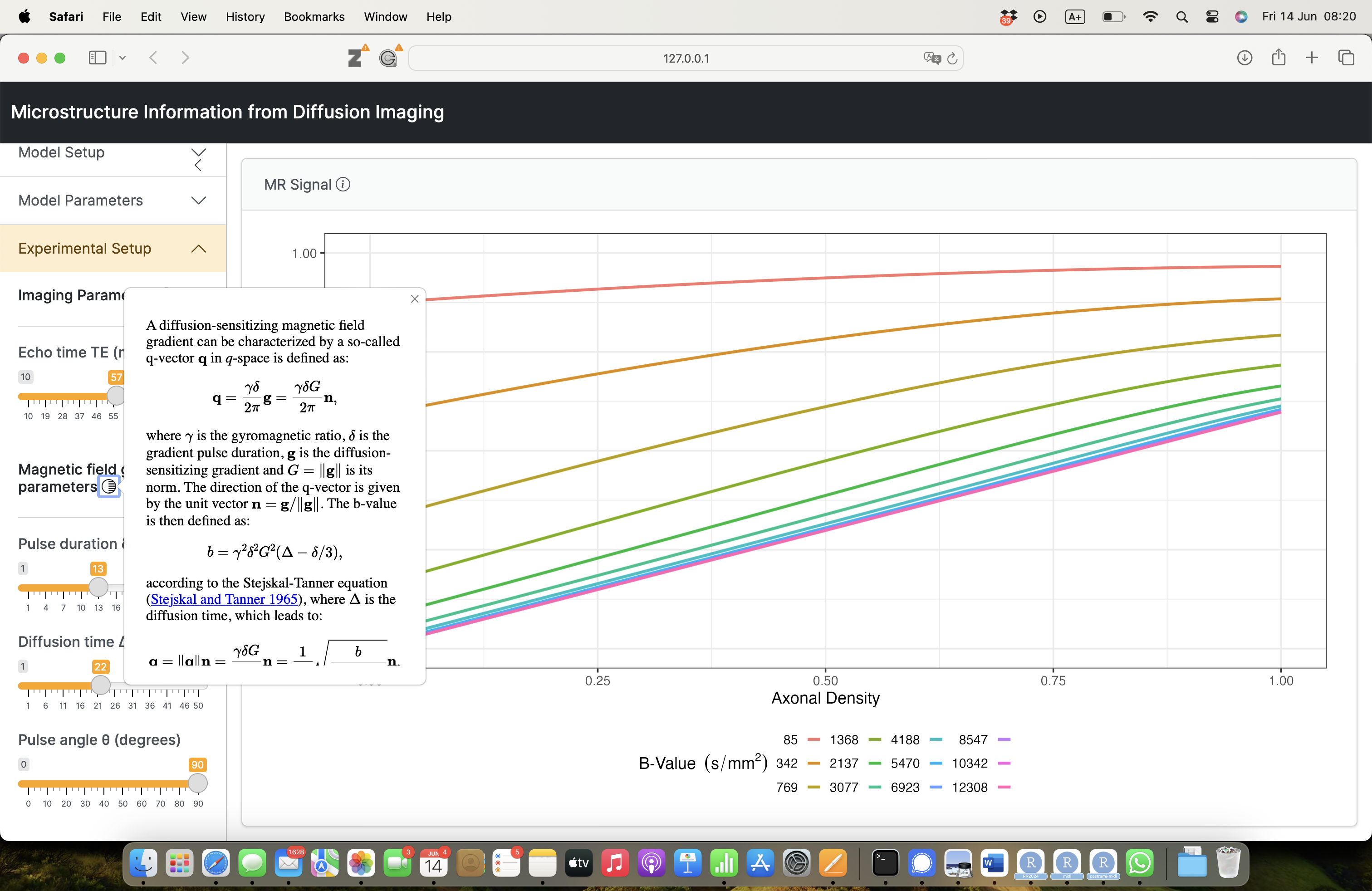
Imaging parameters
\[ \mathbf{q} = \frac{\gamma \delta G}{2 \pi} \mathbf{n} \]
- \(\gamma\): gyromagnetic ratio;
- \(\delta\): gradient duration;
- \(G\): gradient amplitude;
- \(\mathbf{n}\): gradient direction.
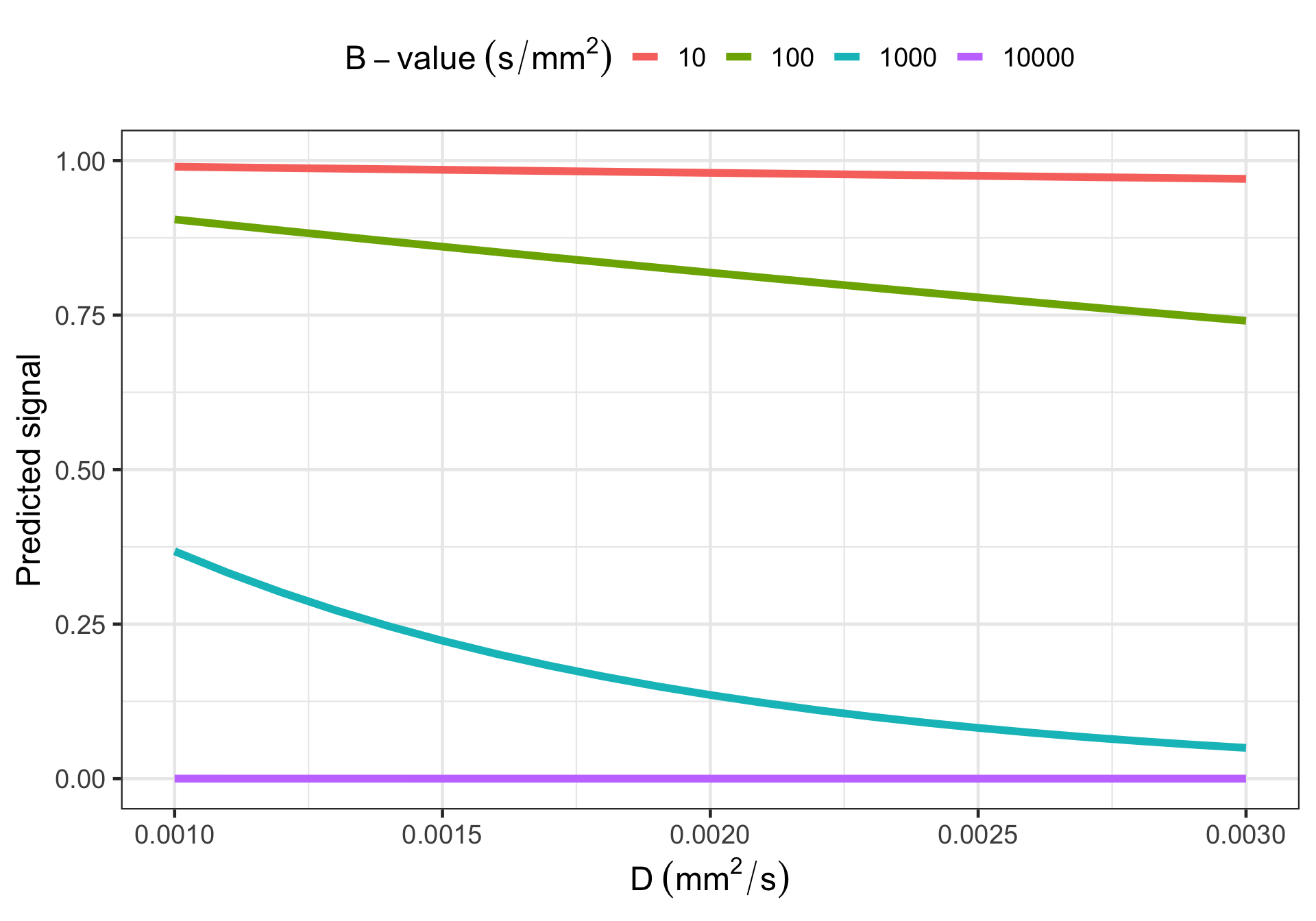
Sensitivity to microstructure
Freely diffusing water:
\[ S(\mathbf{q}) = S_0 \exp \left( - b D \right) \]
with \(b = \gamma^2 \delta^2 G^2 \left( \Delta - \frac{\delta}{3} \right)\).
midi – a tool powered by R and Shiny
Overview
Objective
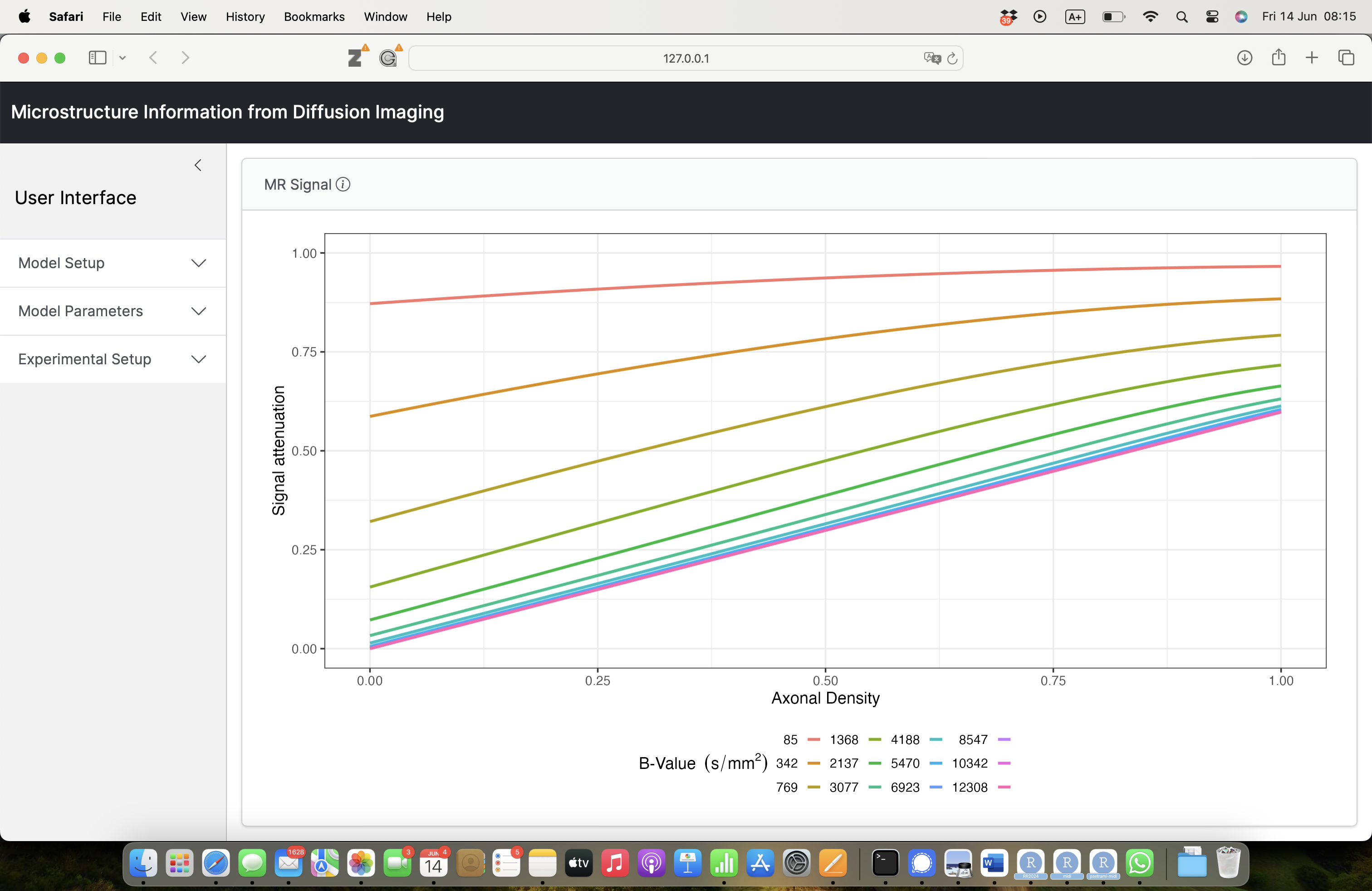
- Shiny for user-friendly interface to simulate MR signals from microstructure models;
- Design best experimental protocols for accurate estimation of a given set of microstructural parameters of interest.
How does it work ?
- choose a set of experimental conditions;
- choose a multi-compartment model to describe microstructure;
- pick a microstructural parameter of interest;
- fix the other model parameters (with sensible defaults provided by the app);
- plot the MR signal wrt the microstructural parameter of interest.
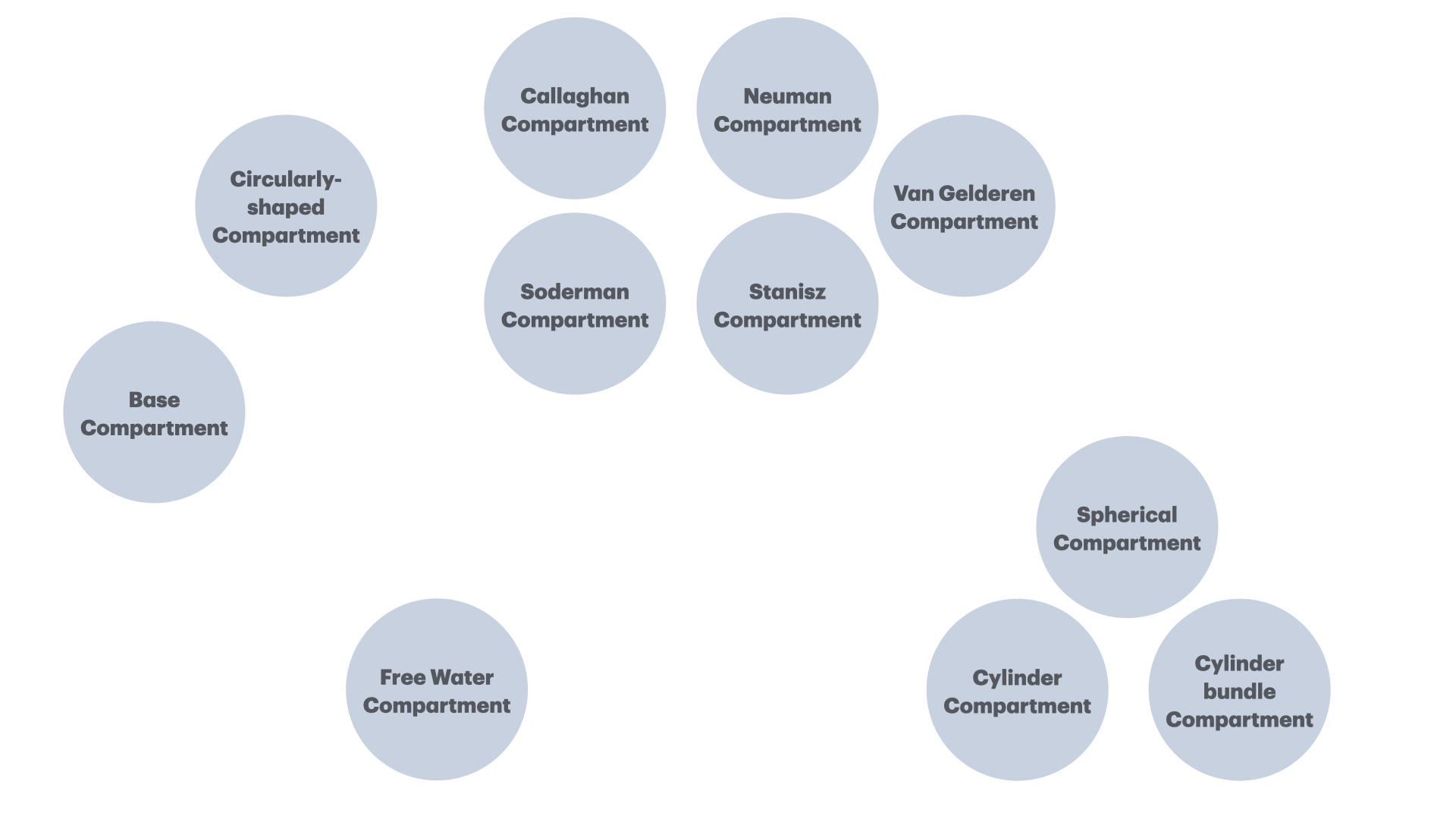
The midi package
An R6-based package
#' Base compartment class
#'
#' @description The base class for compartment models.
#'
#' @keywords internal
BaseCompartment <- R6::R6Class(
"BaseCompartment",
public = list(
#' @description Computes the signal attenuation predicted by the model.
#'
#' @param small_delta A numeric value specifying the duration of the
#' gradient pulse in milliseconds.
#' @param big_delta A numeric value specifying the duration between the
#' gradient pulses in milliseconds.
#' @param G A numeric value specifying the strength of the gradient in
#' mT.\eqn{\mu}m\eqn{^{-1}}.
#' @param direction A numeric vector specifying the direction of the
#' gradient. Defaults to `c(0, 0, 1)`.
#' @param echo_time A numeric value specifying the echo time in
#' milliseconds.
#' @param n_max An integer value specifying the maximum order of the Bessel
#' function. Defaults to `20L`.
#' @param m_max An integer value specifying the maximum number of extrema
#' for the Bessel function. Defaults to `50L`.
#'
#' @return A numeric value storing the predicted signal attenuation.
#'
#' @examples
#' freeComp <- FreeCompartment$new()
#' freeComp$get_signal(small_delta = 30, big_delta = 30, G = 0.040)
#'
#' sphereComp <- SphereCompartment$new()
#' sphereComp$get_signal(small_delta = 30, big_delta = 30, G = 0.040)
#'
#' sodermanComp <- SodermanCompartment$new()
#' sodermanComp$get_signal(small_delta = 30, big_delta = 30, G = 0.040)
#'
#' staniszComp <- StaniszCompartment$new()
#' staniszComp$get_signal(small_delta = 30, big_delta = 30, G = 0.040)
#'
#' neumanComp <- NeumanCompartment$new()
#' neumanComp$get_signal(
#' small_delta = 30, big_delta = 30, G = 0.040,
#' echo_time = 40
#' )
#'
#' callaghanComp <- CallaghanCompartment$new()
#' callaghanComp$get_signal(small_delta = 30, big_delta = 30, G = 0.040)
#'
#' vanGelderenComp <- VanGelderenCompartment$new()
#' vanGelderenComp$get_signal(small_delta = 30, big_delta = 30, G = 0.040)
get_signal = function(small_delta, big_delta, G,
direction = c(0, 0, 1),
echo_time = NULL,
n_max = 20L,
m_max = 50L) {
private$compute_signal(
small_delta = small_delta,
big_delta = big_delta,
G = G,
direction = direction,
echo_time = echo_time,
n_max = n_max,
m_max = m_max
)
},
#' @description Returns the names of the compartment parameters
#'
#' @return A character vector storing the names of the compartment
#' parameters.
#'
#' @examples
#' freeComp <- FreeCompartment$new()
#' freeComp$get_parameter_names()
get_parameter_names = function() {
names(private$parameters())
},
#' @description Returns the values of the compartment parameters
#'
#' @return A numeric vector storing the values of the compartment
#' parameters.
#'
#' @examples
#' freeComp <- FreeCompartment$new()
#' freeComp$get_parameters()
get_parameters = function() {
private$parameters()
}
)
)Should faciliate implementation of new compartment models.
Class hierarchy
The midi Shiny application
V1 powered by Quarto dashboard
- I learned a lot about Quarto dashboards
- The design is not very appealing
- The engine behind the scene is bslib
A better app design
\[ \scriptsize{ \color{cornflowerblue}{S}(\color{salmon}{\mathbf{q}}; \Theta, \mathbf{f}) = \color{lightgreen}{f_\mathrm{free}} \cdot S_\mathrm{free}(\color{salmon}{\mathbf{q}}) + \color{lightgreen}{f_\mathrm{sphere}} \cdot \color{gold}{S_\mathrm{sphere}}(\color{salmon}{\mathbf{q}}; \color{lightgreen}{\Theta_\mathrm{sphere}}) \\ + \color{gold}{f_\mathrm{cylbundle}} \cdot \color{gold}{S_\mathrm{cylbundle}}(\color{salmon}{\mathbf{q}}; \color{lightgreen}{\Theta_\mathrm{cylbundle})}, \quad f_\mathrm{free} + f_\mathrm{sphere} + f_\mathrm{cylbundle} = 1 } \]
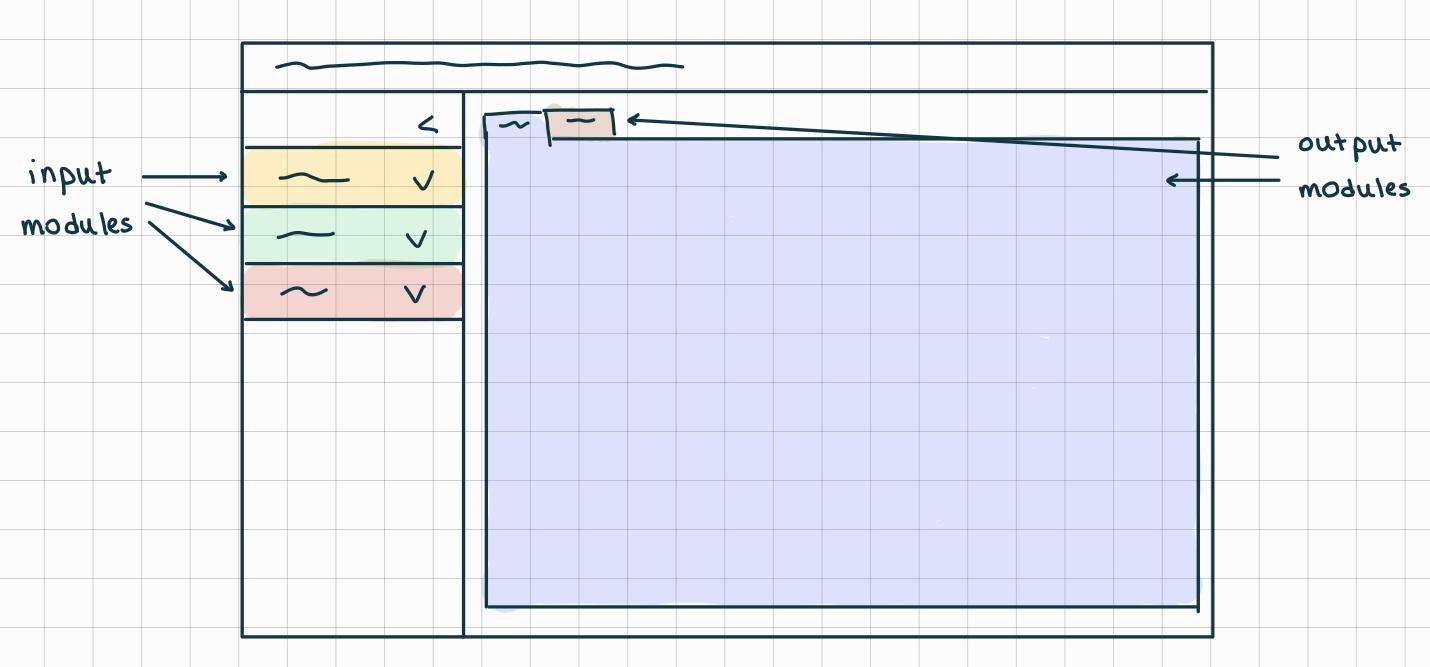
V2 powered by shiny and bslib
V2 powered by shiny and bslib
V2 powered by shiny and bslib
V2 powered by shiny and bslib
V2 powered by shiny and bslib
V2 powered by shiny and bslib
V2 powered by shiny and bslib
rhinoisation (1/2)
rhinoisation (2/2)
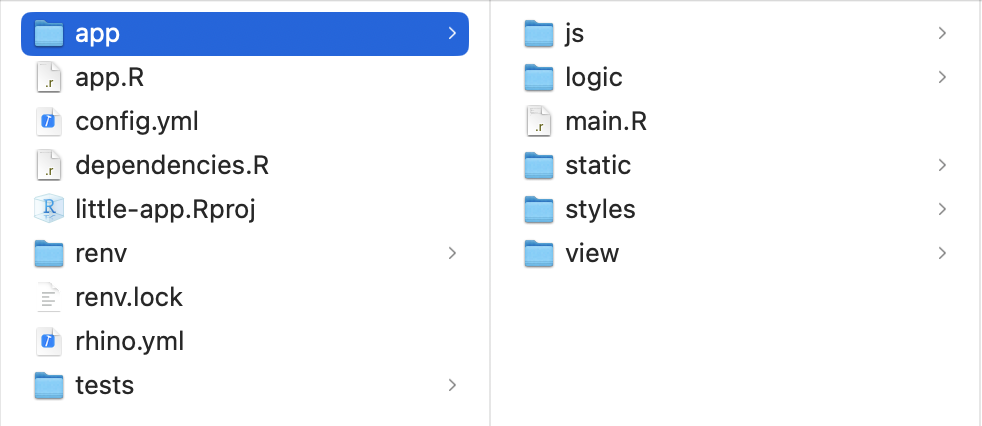
app directory
main.R: app source code;js: javascript code;logic: R objects other than UI/Server;static: images or other static files;styles:scsscode;view: modules.
Root files
app.R: contains onlyrhino::app()to launch the application;dependencies.R: lists all dependencies;renv/renv.lock: handles dependency versions;config.yml/rhino.yml: configuration files;tests: folder containing files for testing.
References

Rencontres R 2024 – Vannes – https://astamm.github.io – aymeric.stamm@cnrs.fr